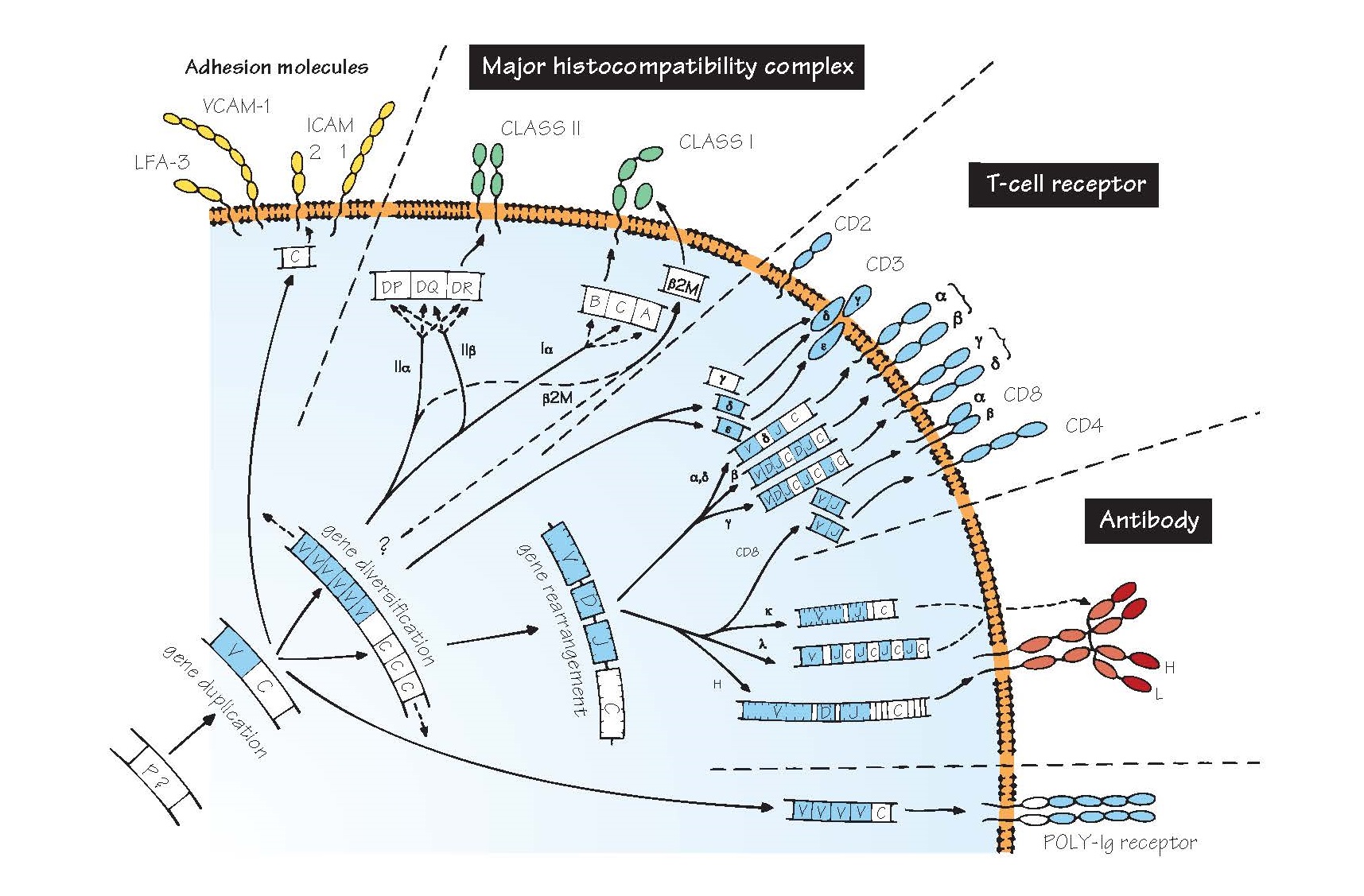
Evolution Of Recognition Molecules: The Immunoglobulin Super Family.
At this point it may be worth re-emphasizing the difference between
‘innate’ and ‘adaptive’ immunity, which
lies essentially in the degree of discrimination of the respective
recognition systems.
Innate immune recognition, e.g. by
phagocytic cells, NK cells or the alternative complement pathway, uses a
limited number of different receptors (more are being discovered all the time,
but there are probably only a few dozen in total), which have evolved to
recognize directly the most important classes of pathogen (see Figs 3 and 5).
Recognition by lymphocytes,
the fundamental cells of adaptive immunity, is quite another matter. An
enormous range of foreign substances can be individually distinguished and the
appropriate response set in motion. This is only possible because of the
evolution of three sets of cell-surface receptors, each showing
extensive heterogeneity, namely the antibody molecule, the T-cell
receptor and the molecules of the major histocompatibility complex (MHC).
Thanks to molecular biology, the fascinating discovery was made that all these
receptors share enough sequences, at both the gene (DNA) and protein (amino acid) level, to make it clear that they
have evolved from a single
precursor, presumably a primitive recognition molecule of some kind (see Figs 3
and 46). The three-dimensional structure of all these receptors which was
obtained more recently using X-ray crystallography has confirmed this close
relationship.
Because antibody was the first of
these genetic systems to be identified, they are often collectively referred to
as the immunoglobulin gene superfamily, which contains other related
molecules too, some with immunological functions, some without. What they all
share is a structure based on a number of folded sequences about 110 amino
acids long and featuring β-pleated sheets, called domains (shown in the
figure as oval loops protruding from the cell membrane).
Much work is still needed to fill
in the evolutionary gaps, and the figure can only give an impression of what
the relationships between this remarkable family of molecules may have been.
Their present-day structure and function are considered in more detail in the
following four figures.
P? The precursor gene from which the immunoglobulin superfamily is presumed to have evolved. It is believed
that the key to the evolutionary success of the characteristic immunoglobulin domain
is its extreme resistance to chemical or physical destruction. The gene has
not been identified in any existing species, but may well have coded for a molecule
that mediated cell–cell recognition. Alternative mechanisms for generating very
diverse families of recognition molecules have been discovered very recently in
several invertebrates and primitive vertebrates, some of which seem to be based
on the leucine-rich repeat (LRR) protein domain instead of the immunoglobulin
domain (see Fig. 5).
V, C A vital early step seems to have been the
duplication of this gene into two, one of which became the parent of all
present-day variable
(V) genes and the other of constant
(C) genes. In the figure, the genes and polypeptides with sufficient
homology to be considered part of the V gene family are shown in blue.
Subsequent further duplications, with diversification among different V and C
genes, led ultimately to the large variety of present-day domains.
Major histocompatibility complex
The genes shown are those
found in humans, also known as HLA (human leucocyte antigen) genes.
Interactions between MHC molecules and T-cell receptors are vital to all
adaptive immune responses. Further details are shown in Fig. 11.
β2M β2-Microglobulin, which combines with class I chains to complete the
four-domain molecule.
Gene rearrangement A process found only in T and B cells, through
which an enormous degree of receptor diversity is generated by bringing
together one V gene and one J gene (and one D gene in the case of IgH chains),
each from a set containing from 2 to over 100. The joins between the segments
are imprecise, leading to millions of possible receptors (see Figs 12 and 13).
This unique process of chromosomal gene rearrangement is brought about by
enzymes called recombinases.
T-cell receptor (TCR) A complex of T-cell surface molecules,
including TCR α plus β, or γ plus δ chains, CD3 and CD4 or CD8, depending on
the type of T cell. Together these form a unit that enables the T cell to
recognize a specific antigen plus a particular MHC molecule, to become
activated and to carry out its function (help, cytotoxicity, etc.; for more
details see Fig. 12).
Antibody The antibody or immunoglobulin molecule plays
the part of cell-surface receptor on B lymphocytes as well as being secreted in
vast amounts by activated B cells to give rise to serum antibody, a vital part
of defence against infectious organisms. The domains are fairly similar to
those of the TCR α and β chains, but assembled in a different way, with two
four-domain heavy (H) chains bonded to two two-domain light (L) chains (see
Figs 13 and 14).
Note that the process of
diversification in the genes for the various chains has not always proceeded in
the same way. For example, mammalian heavy and light (κ) chains have all their
J genes together, between V and C, while light (λ) chains have repeated J–C
segments and sharks have the whole V–D–J–C segment duplicated, a considerably
less efficient arrangement for generating the maximum diversity.
Costimulatory molecules T-cell proliferation and cytokine release (see
Fig. 21) is governed both by the TCR binding to antigen presented on MHC molecules (see Fig. 18) and by
interactions between cell molecules
on the membrane of T cells and their partners (ligands) on the antigen-presenting cell. Many of these
molecules belong to the immunoglobulin superfamily. Some (e.g. CD28 on the T
cell and CD80 or CD86 on the antigen-presenting cell) increase the activation
of the T cell (see also Fig. 12). Others, e.g. CTLA4 and PD1 on the T cell, and
their ligands on the antigen-presenting cell inhibit T-cell activation, and act
to limit or switch off the immune response. Several viruses seem to be able to
increase expression of these negative regulators in order to escape being killed
by the immune system.
Poly-Ig receptor A molecule found on some epithelial cells that
helps to transport antibody into secretions such as mucus. Many other molecules
contain the characteristic immunoglobulin superfamily domain structure,
including some Fc receptors, adhesion molecules (see below) and receptors for
growth factors and cytokines. The common feature seems to be an involvement in
cell–cell interactions, with the ‘breakaway’ immunoglobulin molecule the
exception rather than the rule.
Killer inhibitory receptors
(KIR) Immunoglobulin-family
receptors are found on NK cells (see Fig. 15). They recognize MHC molecules on
target cells and send negative signals to NK cells that inhibit their
activation, and hence prevent killing of targets. NK cells are therefore active
only against cells that have lost MHC expression, either as a result of
infection (e.g. by viruses) or as a result of malignant trans- formation (i.e.
cancer cells). Some NK cells also express other negative receptors that belong
to a different structural family of molecules known as C-lectins. An inhibitory
signalling motif (known as an immunoreceptor tyrosine-based inhibitory motif,
ITIM) on KIR cytoplasmic tails has an important role in the signal transduction
process.
Adhesion molecules A large range of surface molecules help to hold
cells together and facilitate cell–cell interactions or binding to blood vessel
walls. Many of these are involved in regulating inflammation (see Fig. 7) and
attempts to block them for therapeutic purposes are being actively explored.
Some of these, as shown in the figure, belong to the immunoglobulin
superfamily, and they usually bind to one or a small number of corresponding
‘ligands’. Some examples of pairs of molecules important in adhesion are shown
in the table. Many of these molecules have both ‘common’ names and CD numbers
(see Appendix III).