Genetics of Antibody Diversity and Function
Antibody genes are produced by
somatic recombination
The
immunoglobulin repertoire is encoded for by multiple germline gene segments
that undergo somatic diversification in developing B‐cells. Hence, although the
basic components needed to generate an immunoglobulin repertoire are inherited,
an individual’s mature antibody repertoire is essentially formed during their
lifetime by alteration of the inherited germline genes. The first evidence that
immunoglobulin genes rearrange by somatic recombination was reported
by Hozumi and Tonegawa in 1976 (Milestone 3.2). Because somatic recombination
involves rearrangement of DNA in somatic rather than gamete cells, the newly
recombined genes are not inherited. As a result, the primary immunoglobulin
repertoire will differ slightly from one individual to the next, and will be
further modified during an individual’s lifetime by their exposure to different
antigens.
Milestone 3.2
The 1987 Nobel Prize in Physiology or Medicine
Susumu Tonegawa was awarded the 1987 Nobel Prize in Physiology or
Medicine for “his discovery of the genetic principle for generation of antibody
diversity.” In his 1976 paper, Tonegawa used Southern blot analysis of
restriction enzyme digested DNA from lymphoid and nonlymphoid cells to show
that the immunoglobulin variable and constant genes are distant from each other
in the germline genome. Embryo DNA showed two components when hybridized to RNA
probes specific for: (i) both variable and constant regions and (ii) only the
constant region, whereas both probes localized to a single band when hybridized
to DNA from an antibody‐ producing plasmacytoma cell. He proposed that the
differential hybridization patterns could be explained if the variable and
constant genes were distant from each other in germline DNA, but came together
to encode the complete immunoglobulin gene during lymphocyte differentiation.
The immunoglobulin variable gene
segments and loci
The variable
light and heavy chain loci in humans contain multiple gene segments, which are
joined, using somatic recombination, to produce the final V region exon. The
human heavy chain variable region is constructed from the joining of three gene
segments, V (variable), D (diversity),
and J (joining), whereas the light chain variable
gene is constructed by the joining of two gene segments, V and J. There are
multiple V, D, and J segments at the heavy chain and light chain loci, as
illustrated in Figure 3.20.
The human VH
genes have been mapped to chromosome 14, although orphan IgH genes have also
been identified on chromosomes 15 and 16. The human VH locus, as for
other antibody gene segments, is highly polymorphic, and has likely evolved
through the repeated duplication, deletion, and recom bination of DNA.
Polymorphisms found within the germline repertoire are due to the insertion or
deletion of gene segments or the occurrence of different alleles of the same
segment. A number of pseudogenes, ranging from those that are more conserved
and contain a few point mutations to those that are more divergent with
extensive mutations, are also present in immunoglobulin loci. There are
approximately 100 human VH genes, which can be grouped into seven
families based on sequence homology. Members of a given family show approxi
mately 80% sequence homology at the nucleotide level. The functional heavy
chain repertoire is formed from approxi mately 40 functional VH genes, 23 DH
genes and 6 JH genes. The human lambda locus maps to chromosome 22, with
approximately 30 functional Vλ genes and 5 functional Jλ gene segments. The Vλ
genes can be grouped into 10 families. The human kappa locus on chromosome 2 is
composed of a total of approximately 40 functional Vk genes and 5 functional Jk
genes. However, the kappa locus contains a large duplication of most of the Vk
genes, and most of the Vk genes in this distal cluster, although functional,
are seldom used. The numbers of V genes vary between individuals as a result of
polymorphisms.
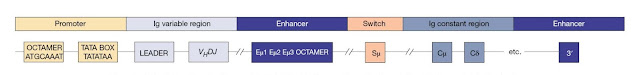
The
immunoglobulin loci also contain regulatory elements (Figure 3.21) including
enhancers at the 3′ end of each locus and also in between the J and C regions
(intronic enhancer) of the IGH and IGK loci. Both 3′ and intronic enhancers are
important for V(D)J recombination, whereas the 3′ enhancers are more important
for the efficient transcription of rearranged Ig genes. Some Ig loci have
additional enhancer elements. Each Ig V gene has its own leader sequence and a
simple promoter that contains a conserved octamer motif and a TATA box.
V(D)J recombination and combinatorial diversity
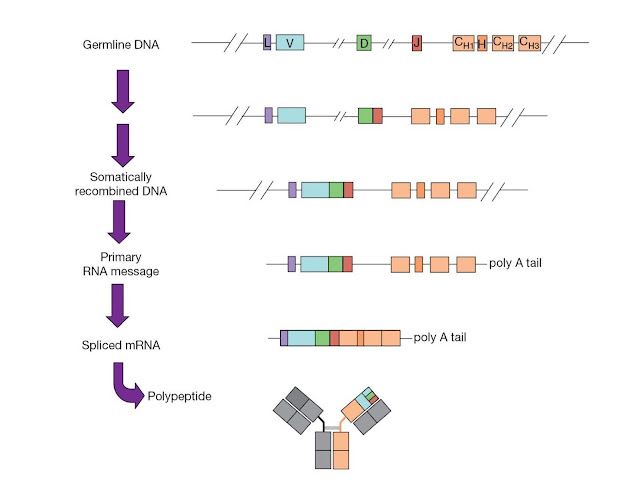
The joining
of these gene segments, illustrated in Figure 3.22, is known as V(D)J
recombination. V(D)J recombination is a highly regulated and ordered
event. The light chain exon is constructed from a single V‐to‐J gene segment
join. However, at the heavy chain locus, a D segment is first joined to a J
segment, and then the V segment is joined to the combined DJ sequence. The
rearranged DNA is transcribed, the RNA transcript is spliced to bring together
the V region exon and the C region exon, and lastly the spliced mRNA is
translated to produce the final immunoglobulin protein.
Numerous
unique immunoglobulin genes can be made by joining different combinations of
the V, D, and J segments at the heavy and light chain loci. The creation of
diversity in the immunoglobulin repertoire through this joining of various gene
segments is known as combinatorial diversity. Additional
diversity is created by the pairing of different heavy chains with different
lambda or kappa light chains. For example, the potential heavy chain repertoire
is very approximately 40 VH × 23 DH × 6 JH =
5500 different combinations. Similarly, there are very approximately 150 (30
Vλ× 5 Jλ) and 200 (40 Vk × 5 Jk) different combinations, for a total of 350
light chain combinations. If we consider that each heavy chain could
potentially pair with each light chain, then the diversity of the
immunoglobulin repertoire would be quite large, on the order of 2 million
possible combinations. However, V genes rearrange at very different
frequencies, so there is enormous variation in the likelihood of different
combinations. Additional diversity is also generated during gene segment
recombination and by somatic hypermutation, as explained in the following
sections. In this manner, although the number of germline gene segments appears
limited in size, an incredibly diverse immunoglobulin repertoire can be
generated.
Recombination signal sequences
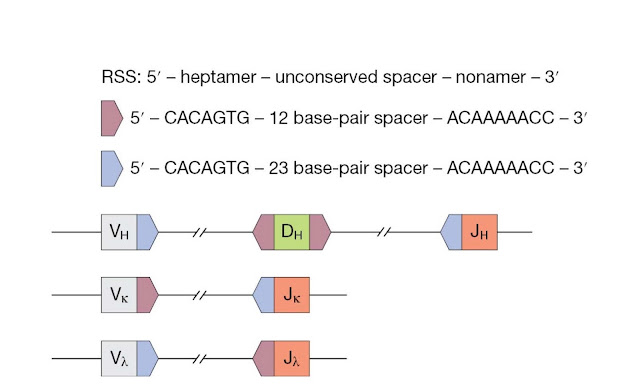
The recombination
signal sequence (RSS) helps to guide recombination between appropriate
gene segments. The RSS (Figure 3.23) is a noncoding sequence that flanks coding
gene segments. It is made up of a conserved heptamer and nonamer sequences,
which are separated by an unconserved 12‐ or 23‐nucleotide spacer. Efficient
recombination occurs between segments with a 12‐nucleotide spacer and a
23‐nucleotide spacer. This “12/23” rule helps make certain that
appropriate gene segments are joined together.
At the VH
locus, the V and J segments are flanked by RSSs with a 23‐nucleotide spacer,
whereas the D segments are flanked by RSSs with a 12‐nucleotide spacer. At
light chain loci, the Vk segments are flanked by RSSs with 12‐nucleotide
spacers, Jk segments are flanked by RSSs with 23‐nucleotide spacers, and this
arrangement is reversed in the lambda locus.
The recombinase machinery
The V(D)J
recombinase is a complex of enzymes that mediates somatic recombination of
immunoglobulin gene segments (Figure 3.24). The gene products of recombination‐activating
genes 1 and 2 (RAG‐1 and RAG‐2) are lymphocyte‐specific enzymes essential for
V(D)J recombination. In the initial steps of V(D)J recombination, the RAG
complex binds the recombination signal sequences and, in association with high mobility
group (HMG) proteins that are involved in DNA bending, the two recombination
signal sequences are brought together. In contrast to the lymphoid‐specific RAG
enzymes, HMG proteins are ubiquitously expressed.
Next, a
single‐stranded nick is introduced between the 5′‐heptameric end of the
recombination signal sequence and the coding segment. This nick results in a
free 3′ OH group, which attacks the opposite, anti‐parallel DNA strand in a
transesterification reaction. This attack gives rise to a double‐ stranded DNA
break that leads to the formation of covalently sealed hairpins at the two
coding ends and the formation of blunt signal ends. At this stage a
post‐cleavage complex is formed, in which the RAG recombinase remains
associated with the DNA ends.
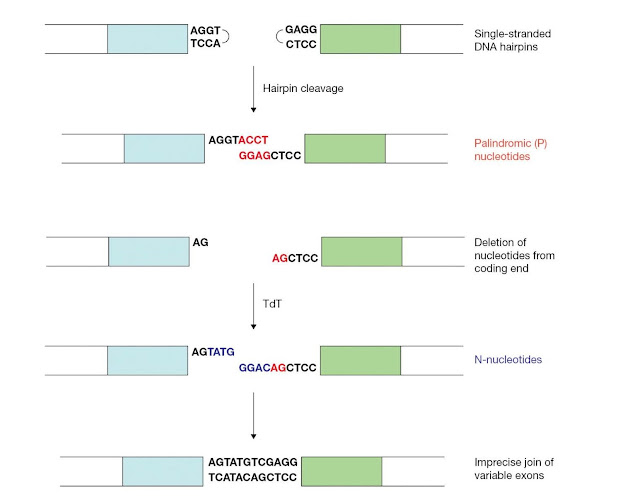
The DNA
break is finally repaired by nonhomologous end‐joining machinery. The
recombination signal sequences are joined precisely to generate the signal
joint. By contrast, nucleotides can be lost or added during repair of the
coding ends (Figure 3.25). Junctional diversity is the
diversification of variable region exons due to this imprecise joining of the
coding ends.
First, a small number of nucleotides are often deleted from the coding end by an unknown exonuclease. Also, junctional diversity involves the potential addition of two types of nucleotides, P‐nucleotides and N‐nucleotides. The palindromic sequences that result from the asymmetric cleavage and template‐mediated fill‐in of the coding hairpins are referred to as P‐nucleotides. N‐nucleotides are generated by the nontemplated addition of nucleotides to the coding ends, which is mediated by the enzyme terminal deoxynucleotidyl transferase (TdT). Although P‐ and N‐nucleotides and deletion of the coding end and nucleotides serve to greatly diversify the immunoglobulin repertoire, the addition of these nucleotides may, as for other events in antibody gene assembly, result in the genera tion of receptor genes that are out of frame.
First, a small number of nucleotides are often deleted from the coding end by an unknown exonuclease. Also, junctional diversity involves the potential addition of two types of nucleotides, P‐nucleotides and N‐nucleotides. The palindromic sequences that result from the asymmetric cleavage and template‐mediated fill‐in of the coding hairpins are referred to as P‐nucleotides. N‐nucleotides are generated by the nontemplated addition of nucleotides to the coding ends, which is mediated by the enzyme terminal deoxynucleotidyl transferase (TdT). Although P‐ and N‐nucleotides and deletion of the coding end and nucleotides serve to greatly diversify the immunoglobulin repertoire, the addition of these nucleotides may, as for other events in antibody gene assembly, result in the genera tion of receptor genes that are out of frame.
Similar to
the RAG recombinase complex, the DNA repair machinery works as a
protein complex. However, unlike the RAG recombinase, the nonhomologous
end‐joining proteins are ubiquitously expressed. In the first steps of DNA
repair, the Ku70 and Ku80 proteins form a heterodimer that binds the broken DNA
ends. The Ku complex recruits the catalytic subunit of DNA‐dependent protein
kinase (DNA‐PKcs), a serine‐threonine protein kinase. The activated DNA‐PKcs
then recruits and phosphorylates XRCC4 and Artemis. Artemis is an endonuclease
that opens the hairpin coding ends. Finally, DNA ligase IV binds XRCC4 to form
an end‐ligation complex, and this complex mediates the final ligation and
fill‐in steps needed to form the coding and signal joints.
Regulating V(D) J recombination
V(D)J
recombination and the recombinase machinery must be carefully regulated to
avoid wreaking havoc on the cellular genome. For instance, aberrant V(D)J
recombination is implicated in certain B‐cell lymphomas. V(D)J recombination is
largely regulated by controlling expression of the recombination machinery and
the accessibility of gene segments and nearby enhancers and promoters. As
previously mentioned, RAG‐1 and RAG‐2 activity is specific to lymphoid cells,
and further regulation is imposed by downregulating RAG activity during
appropriate stages of B‐cell development. Differential accessibility of gene
segments to the recombinase machinery, which can be achieved by altering
chromatin structure, also plays a role in making certain that appropriate gene
segments are recombined in an appropriate order. Cis‐acting transcrip
tional control elements, such as enhancers and promoters, also help regulate
recombination. Although it is not a hard and fast rule, transcription from
certain regulatory elements seems to correlate with rearrangement of the
adjacent genes. This sterile, or nonproductive, transcription
may somehow help target required proteins or modulate gene
accessibility. Finally, in addition to directing recombination between
appropriate gene segments, the precise sequences of the RSS itself, as well as
the sequences of the gene segments themselves, can influence the efficiency of
the recombination reaction.
Somatic hypermutation
Following
antigen activation, the variable regions of immunoglobulin heavy and light
chains are further diversified by somatic hypermutation. Somatic
hypermutation involves the introduction of nontemplated point mutations
into V regions of rapidly proliferating B‐cells in the germinal centers of
lymphoid follicles. Antigen‐driven somatic hypermutation of variable
immunoglobulin genes can result in an increase in binding affinity of the
B‐cell receptor for its cognate ligand. As B‐cells with higher affinity
immunoglobulins can more successfully compete for limited amounts of antigen
present, an increase in the average affinity of the antibodies produced during
an immune response is observed. This increase in the average affinity of
immunoglobulins is known as affinity maturation.
Somatic
hypermutation occurs at a high rate, thought to be on the order of about 1 ×
10−3 mutations per base‐pair per generation, which is approximately 106 times
higher than the mutation rate of cellular housekeeping genes. There is a bias
for transition mutations, and the “mutation hotspots” in variable regions map
to RGWY motifs (R = purine, Y =
pyrimidine, W= A or T). The exact mechanisms by which mutations are introduced
and preferentially targeted to appropriate V regions, while constant regions of
the immunoglobulin loci remain protected, is not clearly understood and is the
subject of current research. Transcription through the target V region seems
required, but is not sufficient, for somatic hypermutation. Additionally, the
enzyme activation‐induced cytidine deaminase (AID)
has been demonstrated to be essential for both somatic hypermutation and class
switch recombination.
AID is a
cytidine deaminase capable of carrying out targeted deamination of C to U, and
shows strong homology with the RNA‐editing enzyme APOBEC‐1. It appears that AID
directly deaminates DNA to produce U : G mismatches. The exact mechanism by
which AID can differentially regulate somatic hypermutation and class switch
recombination is currently being studied, and may depend on interactions of
specific cofactors with specific domains of AID.
Therefore,
diversity within the immunoglobulin repertoire is generated by: (i) the
combinatorial joining of gene segments; (ii) junctional diversity; (iii)
combinatorial pairing of heavy and light chains; and (iv) somatic hypermutation
of V regions.
Gene conversion and repertoire diversification
Although
mice and humans use combinatorial and junctional diversity as a mechanism to
generate a diverse repertoire, in many species, including birds, cattle, swine,
sheep, horses, and rabbits, V(D)J recombination results in assembly and expres
sion of a single functional gene. Repertoire diversification is then achieved
by gene conversion, a process in which pseudo‐ V genes are used
as templates to be copied into the assembled variable region exon. Further
diversification may be achieved by somatic hypermutation.
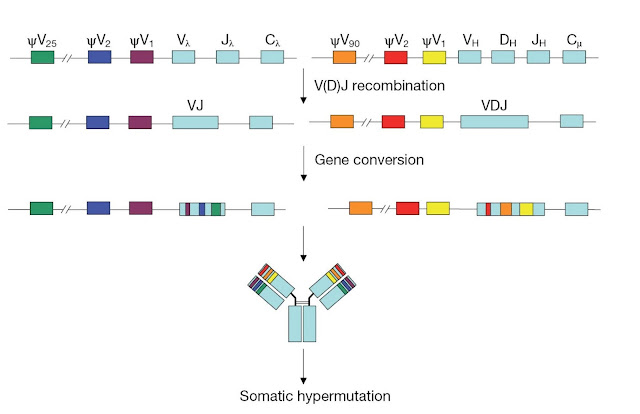
The process
of gene conversion was originally identified in chickens, in which immature
B‐cells have the same variable region exon. During B‐cell development in the
bursa of Fabricius, rapidly proliferating B‐cells undergo gene conversion to
diversify the immunoglobulin repertoire (Figure 3.26). Stretches of sequences
from germline variable region pseudogenes, located upstream of the functional V
genes, are introduced into the VL and VH regions. This process takes place in
the ileal Peyer’s patches of cattle, swine, and horses, and in the appendix of
rabbits. These gut‐associated lymphoid tissues are the mammalian equivalent of
the bursa in these species.
Class switch recombination
Antigen‐stimulated
IgM expressing B‐cells in germinal centers of secondary lymphoid organs, such
as the spleen and lymph nodes, undergo class switch recombination. Class
switch recombination (CSR) allows the IgH constant region
exon of a given antibody to be exchanged for an alternative exon, giv ing rise
to the expression of antibodies with the same antigen specificity but of
differing isotypes, and therefore of differing effector functions as described
earlier. CSR occurs through a deletional DNA recombination event at the IgH
locus (Figure 3.27), which has been extensively studied in mice. Constant
region exons for IgD, IgG, IgE, and IgA isotypes are located downstream of the
IgM (Cμ) exon, and CSR occurs between switch or S regions.
S regions are repetitive sequences, which are often G‐rich on the nontemplate
strand, that are found upstream of each CH exon except Cδ. Breaks are
introduced into the DNA of two S regions and fusion of the S regions leads to a
rearranged CH locus, in which the variable exon is joined to an exon for a new
constant region. The DNA between the two switch regions is excised and forms an
episomal circle. Finally, alternative splicing of the primary RNA transcript
generated from the rearranged DNA gives rise to either membrane‐bound or
secreted forms of the immunoglobulin.
Prior to
recombination between switch regions, transcription is initiated from a
promoter found upstream of an exon that precedes all CH genes capable of
undergoing CSR, the intervening (I) exon. These germline transcripts include I,
S, and C region exons, and do not appear to code for any functional protein.
However, this germline transcription is required, although not sufficient, to
stimulate CSR. The precise mecha nism responsible for CSR is the subject of
current study, but work indicates that AID, described previously to be involved
in somatic hypermutation, helps mediate CSR, along with some components of the
nonhomologous end‐joining pathway and several other DNA repair pathways. The
joining of S regions may be mediated by association with transcriptional promoters,
enhancers, chromatin factors, DNA repair proteins, AID‐associated factors, or
by interactions involving S region sequences themselves.