The Major
Histocompatibility Complex (MHC)
Molecules
within this complex were originally defined by their ability to provoke
vigorous rejection of grafts exchanged between different members of a species
(Milestone 4.2). We have already referred to the necessity for antigens to be
associated with class I or class II MHC molecules in order that they may be
recognized by T‐lymphocytes (Figure 4.8). How antigenic peptides are processed
and selected for presentation within MHC molecules and how the TCR sees this
complex are discussed in detail in Chapter 5, but let us run through the major
points briefly here so that reader will appreciate why these molecules are of
huge importance within the immune system.
MHC
molecules assemble within the cell, where they associate with short peptide
fragments derived either from proteins being made by the cell (MHC class I
molecules bind to peptides derived from proteins being synthesized within the
cell) or proteins that have been internalized by the cell through phagocytosis
or pinocytosis (MHC class II molecules bind to peptides derived from proteins
made external to the cell). There are some exceptions to these general rules,
which we deal with in Chapter 5. We have already made the analogy that this
process represents a type of “quality control” checking system where a fraction
of proteins present in the cell at any given moment are presented to T‐cells
for inspection to ensure that none of these is derived from nonself. Of course,
if a cell happens to harbor a nonself peptide, we want the immune system to
know about this as quickly as possible, so that the appropri ate course of
action can be taken. Thus, MHC class I molecules display peptides that are
either self, or that are being made by an intracellular virus or bacterium. MHC
class II molecules display peptides that are either extracellular self proteins
or proteins being made by extracellular microorganisms. The whole point is to
enable a T‐cell to inspect what is going on, antigenically speaking, within the
cell.
As we shall
see, MHC class I molecules serve an important role presenting peptides for
inspection by CD8 T‐cells that are mainly preoccupied with finding virally
infected or “abnormal” cells to kill. Should a TCR‐bearing CD8 T‐cell recognize
a class I MHC–peptide combination that is a good “fit” for its TCR, it will
attack and kill that cell. MHC class II molecules, on the other hand, are not
expressed on the general cell population but are restricted to cells of the
immune system, such as DCs, that have an antigen‐presenting function as we
already outlined in Chapter 1. Upon recognition of an appropriate MHC class
II–peptide combination by a CD4 T‐cell, this will result in activation of the
latter and maturation to an effector T‐cell that can give help to B‐cells to
make antibody for example. Although this is an oversimplification, as we will
learn in later chapters, please keep in mind the general idea that MHC class I
and II molecules present peptides to CD8‐ and CD4‐ restricted T‐cells,
respectively, for the purposes of allowing these cells to determine whether
they should become “activated” and differentiate to effector cells. Let us now
look at these molecules in greater detail.
Figure
M4.2.1 Main genetic regions of the major
histocompatibility complex (MHC).

Class I and Class II Molecules Are
Membrane Bound Heterodimers
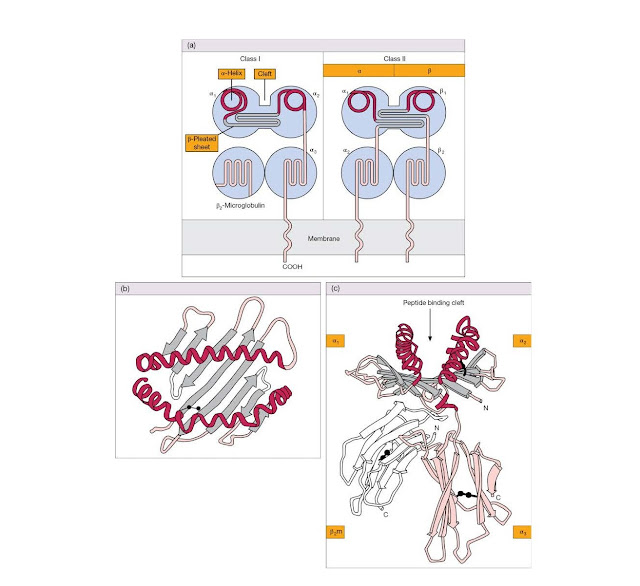
MHC class I
Class I molecules consist of a
heavy polypeptide chain of 44 kDa noncovalently linked to a smaller 12 kDa
polypep tide called β2‐microglobulin. The largest part of
the heavy chain is organized into three globular domains (α1, α2, and α3) that
protrude from the cell surface, a hydrophobic section anchors the molecule in
the membrane, and a short hydrophilic sequence carries the C‐terminus into the
cytoplasm (Figure 4.19).
The solution
of the crystal structure of a human class I molecule provided an exciting leap
forwards in our understanding of MHC function. Both β2‐microglobulin and the α3
region resemble classic Ig domains in their folding pattern (see Figure 4.19c).
However, the α1 and α2 domains, which are most distal to the membrane, form two
extended α‐helices above a floor created by strands held together in a
β‐pleated sheet, the whole forming an undeniable groove (Figure
4.19b,c). The appearance of these domains is so striking, we doubt whether the
reader needs the help of gastronomic analogies such as “two sausages on a
barbecue” to prevent any class I structural amnesia. Another curious feature
emerged. The groove was occupied by a linear molecule, now known to be a
peptide, which had co‐crystallized with the class I protein (Figure 4.20).
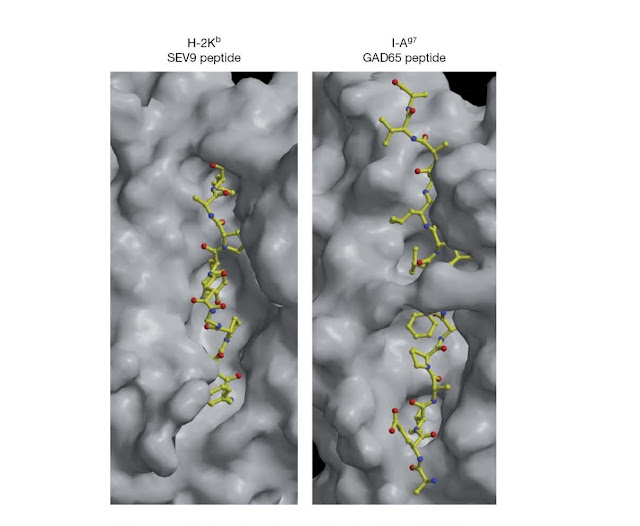
Figure 4.20 Surface
view of mouse class I and class II MHC molecules in complex with peptide.
Surface solvent‐accessible areas of the mouse class I molecule (H‐2Kb) in
complex with a virus‐derived peptide and the mouse class II molecule I‐Ag7 in
complex with an endogenous peptide. The views shown here are similar to that
schematically depicted in Figure 4.19b and look down upon the surface of the
MHC molecules. Note that the peptide‐binding cleft of class I molecules is more
restricted than that of class II molecules, with the result that class
I‐binding peptides are typically shorter than those that bind to class II
molecules. (Source: Dr. Robyn Stanfield and Dr. Ian Wilson, Department of
Molecular Biology, The Scripps Research Institute, La Jolla, California, USA.
Reproduced with permission.)
MHC class II
Class II MHC molecules are also
transmembrane glycoproteins, in this case consisting of α and β polypeptide
chains of molecular weight 34 kDa and 29 kDa, respectively.
There is
considerable sequence homology with class I, and structural studies have shown
that the α2 and β2 domains, the ones nearest to the cell membrane, assume the
characteristic Ig fold, while the α1 and β1 domains mimic the class I α1 and α2
in forming a groove bounded by two α‐helices and a β‐pleated sheet floor
(Figure 4.19a and Figure 4.20).
The
organization of the genes encoding the α chain of the human class II molecule
HLA‐DR and the main regulatory sequences that control their transcription are
shown in Figure 4.21.
Figure 4.21 Genes encoding human HLA‐DR α chain
(darker blue) and their controlling elements (regulatory sequences in light
blue and TATA box promoter in yellow). α1/α2 encode the two extracellular
domains; TM and CYT encode the transmembrane and cytoplasmic segments,
respectively. 3′‐UT represents the 3′‐untranslated sequence. Octamer motifs are
also found in virtually all heavy and light chain immunoglobulin V gene
promoters and in the promoters of other B‐cell‐specific genes such as B29 and
CD20.
MHC class I and class II molecules
are polygenic
Several
different flavors of MHC class I and class II proteins are expressed by most
cells. There are three different class I α‐chain genes, referred to as HLA‐A,
HLA‐B, and HLA‐C in humans and H‐2 K, H‐2D, and H‐2
L in the mouse, which can result in the expression of at least three
different class I proteins in every cell. This number is doubled if an
individual is heterozygous for the class I alleles expressed at
each locus; indeed, this is often the case because of the polymorphic nature
of class I genes, as we shall discuss later in this chapter.
There are
also three different types of MHC class II α‐ and β‐chain genes expressed in
humans, HLA‐DQ, HLA‐ DP, and HLA‐DR, and two pairs in
mice, H2‐A (I‐A) and H2‐E (I‐E). Thus, humans can express a
minimum of three different class II molecules, with this number increasing
significantly when polymorphisms are considered; this is because different α‐
and β‐chain combinations can be gener ated when an individual is heterozygous
for a particular class II gene.
The
different types of class I and class II molecules all exhibit the same basic
structure as depicted in Figure 4.19a and all participate in presenting
peptides to T‐cells but, because of significant differences in their
peptide‐binding grooves, each presents a different range of peptides to
the immune system. This has the highly desirable effect of reducing the
probability that peptides derived from pathogen proteins will fail to be
presented.
Class I and
class II MHC molecules probably evolved from a single ancestral gene that
underwent serial gene duplications, followed by diversification owing to
selective pressure, to generate the different class I and class II genes that
we see today (Figure 4.22). Genes that failed to confer any selective advantage
or that suffered deleterious mutations were either deleted from the genome or
are still present as pseudogenes (genes that fail to express a functional
protein); indeed many pseudogenes are present within the MHC region. This type
of gene evolution pattern has been termed the birth and death model or
the accordion model because of the way in which this gene region expanded and
contracted during evolution.
Several immune response‐related
genes contribute to the remaining class III region of the MHC
A variety of
other genes that congregate within the MHC chromosome region are grouped under
the heading of class III. Broadly, one could say that many are directly or
indirectly related to immune defense functions. A notable cluster involves four
genes coding for complement components, two of which are for the C4 isotypes
C4A and C4B and the other two for C2 and factor B. The cytokines tumor necrosis
factor (TNF, sometimes referred to as TNFα) and lymphotoxin (LTα and LTβ) are
encoded under the class III umbrella, as are three members of the human 70 kDa
heat‐shock proteins. As ever, things do not quite fit into the nice little
boxes we would like to put them in. Even if it were crystal clear where one
region of the MHC ends and another begins (and it isn’t), some genes located in
the middle of the “classical” (see Figure 4.24) class I or II regions should
more correctly be classified as part of the class III cohort. For example, the LMP
and TAP genes concerned with the intracellular processing and
transport of T‐cell epitope peptides are found in the class II region but do
not have the classical class II structure, nor are they expressed on the cell
surface.
The genes of the MHC display
remarkable polymorphism
Unlike the
immunoglobulin system where, as we have seen, variability is achieved in each
individual by a multigenic system, the MHC has evolved in terms
of variability between individuals with a highly polymorphic (literally
“many shaped”) system based on multiple alleles (i.e.,
alternative genes at each locus). This has likely arisen through pathogendriven
selection to form new alleles that may offer increased “fitness” for
the individual; in this context, fitness could mean increased protection from
an infectious organism. The class I and class II genes are the most polymorphic
genes in the human genome; for some of these genes over 600 allelic variants
have been identified (Figure 4.26). This implies that there has been intense
selective pressure on the MHC gene region and that genes within this region are
mutating at rates much faster than those at other gene loci.
As is amply
illustrated in Figure 4.26, class I HLA‐A, ‐B, and ‐C molecules are highly
polymorphic and so are the class II β chains (HLA‐DRβ most, ‐DPβ next, and ‐DQβ
third) and, albeit to a lesser extent than the β chains, the α chains of ‐DP
and ‐DQ. HLA‐DRα and β2‐microglobulin are invariant in structure. The amino
acid changes responsible for this poly morphism are restricted to the α1 and α2
domains of class I and to the α1 and β1 domains of class II. It is of enormous
significance that they occur essentially in the β‐sheet floor and on the inner
surfaces of the α‐helices that line the central cavity (Figure 4.19a) and also
on the upper surfaces of the helices; these are the very surfaces that make
contact with the peptides that these MHC molecules offer up for inspection by
TCRs (Figure 4.20). The nonrandom location at which MHC alleles diverge from
one another is as a result of positive selection over the course of animal
evolution due to host–pathogen interactions. As a consequence of the
polymorphic nature of MHC molecules, the spectrum of peptides bound by these
molecules is highly variable. In Chapter 5 we will explore in greater detail
how peptide interacts with the β‐pleated sheet floor of MHC molecules, as these
interactions dramatically influence the type of peptides that can be presented
by particular molecules. The ongoing drive towards creating new MHC molecules, with
slightly altered peptide‐binding grooves, is akin to a genetic arms
race where the
immune system is constantly trying to keep one step ahead of
its foe. This genetic one‐upmanship has been termed pathogen‐driven
balancing selection because heterozygotes typically have a selective
advantage over homozygotes at a given locus.
The MHC
region represents an outstanding hotspot with mutation rates two orders of
magnitude higher than non‐ MHC loci. These multiple allelic forms can be
generated by a variety of mechanisms: point mutations, recombination,
homologous but unequal crossing over, and gene conversion.
The degree
of sequence homology and an increased occur rence of the dinucleotide motif
5′‐cytosine–guanine‐3′ (to produce what are referred to as CpG islands) seem to
be important for gene conversion, and it has been suggested that this might
involve a DNA‐nicking activity that targets CpG‐rich DNA sequences. MHC genes
that lack these sequences, for example H‐2Ead and HLA‐DRA, do not
appear to undergo gene conversion, whereas those that possess CpG islands act
as either donors (e.g., H‐2Ebb, H‐2Q2k, H‐2Q10b),
acceptors
Figure 4.22 Birth and death model of MHC
evolution. Different major histocompatibility complex (MHC) genes most likely
arose though duplication events that resulted in diversification of the
duplicated genes as a result of selective pressure. Genes that confer no selective
advantage can suffer deleterious mutations resulting in pseudogenes or may be
deleted from the genome altogether. Different environments impose distinct
selective pressures, due to different pathogens for example, resulting in a
high degree of polymorphism within this gene family. MHC polymorphism is seen
primarily within the peptide‐binding regions of MHC class I and class II
molecules.
Gene map of the MHC
The complete
sequence of a human MHC was published at the very end of the last millennium
after a gargantuan collaborative effort involving groups in England, France,
Japan, and the United States. The entire sequence, which represents a composite
of several MHC haplotypes, comprises 224 gene loci. Of the 128 of these genes
that are predicted to be expressed, it is estimated that about 40% of them have
functions related to the immune system. It is not clear why so many immune
response‐related genes are clustered within this relatively small region,
although this phenomenon has also been observed with housekeeping genes that
share related functions. Because the location of a gene within chromatin can
profoundly influence its transcriptional activity, perhaps it has something to
do with ensuring that the genes within this region are expressed at similar
levels. Genes found within con densed regions of chromatin are often expressed
at relatively low levels and in some cases may not be expressed at all. The
region between class II and class I in the human contains 60 or so class III
genes. An overall view of the main clusters of class I, II, and III genes in
the MHC of the mouse and human may be gained from Figure M4.2.1 in Milestone
4.2. More detailed maps of each region are provided in Figure 4.23, Figure
4.24, and Figure 4.25. A number of pseudogenes have been omitted from these
gene maps in the interest of simplicity.
The cell
surface class I molecule, based on a transmembrane chain with three
extracellular domains associated with β2‐microglobulin, has clearly proved to
be a highly useful structure judging by the number of variants on this theme that
have arisen during evolution. It is helpful to subdivide them, first into the classical
class I molecules (sometimes referred to as class Ia), HLA‐A, ‐B, and
‐C in the human and H‐2 K, ‐D, and ‐L in the mouse. These were defined
serologically by the antibodies arising in grafted individuals using methods
developed from Gorer’s pioneering studies (Milestone 4.2).
Other
molecules, sometimes referred to as class Ib, have related structures and are
either encoded within the MHC locus itself (“nonclassical” MHC
molecules, for example the human HLA‐E, ‐F, and ‐G, HFE, MICA and MICB, the
murine H‐2 T, ‐Q, and ‐M), or elsewhere in the genome (“class I
chain‐related,” including the CD1 family and FcRn). Nonclassical MHC
genes are far less polymorphic than the classical MHC, are often invariant, and
many are pseudogenes. Many of these nonclassical MHC class I molecules form
structures that are very similar to class I molecules and have also been found
to either present nonpeptide antigens or canonical (i.e., invariant) peptides
that serve roles in monitoring overall cell stress levels. We will discuss
these non classical MHC molecules in more detail towards the end of this
chapter.
The genes of the MHC display
remarkable polymorphism
Unlike the
immunoglobulin system where, as we have seen, variability is achieved in each
individual by a multigenic system, the MHC has evolved in terms
of variability between individuals with a highly polymorphic (literally
“many shaped”) system based on multiple alleles (i.e., alternative
genes at each locus). This has likely arisen through pathogen‐driven
selection to form new alleles that may offer increased “fitness” for
the individual; in this context, fitness could mean increased protection from
an infectious organism. The class I and class II genes are the most polymorphic
genes in the human genome; for some of these genes over 600 allelic variants have
been identified (Figure 4.26). This implies that there has been intense
selective pressure on the MHC gene region and that genes within this region are
mutating at rates much faster than those at other gene loci.
As is amply
illustrated in Figure 4.26, class I HLA‐A, ‐B, and ‐C molecules are highly
polymorphic and so are the class II β chains (HLA‐DRβ most, ‐DPβ next, and ‐DQβ
third) and, albeit to a lesser extent than the β chains, the α chains of ‐DP
and ‐DQ. HLA‐DRα and β2‐microglobulin are invariant in structure. The amino
acid changes responsible for this poly morphism are restricted to the α1 and α2
domains of class I and to the α1 and β1 domains of class II. It is of enormous
significance that they occur essentially in the β‐sheet floor and on the inner
surfaces of the α‐helices that line the central cavity (Figure 4.19a) and also
on the upper surfaces of the helices; these are the very surfaces that make
contact with the peptides that these MHC molecules offer up for inspection by
TCRs (Figure 4.20). The nonrandom location at which MHC alleles diverge from
one another is as a result of positive selection over the course of animal
evolution due to host–pathogen interactions. As a consequence of the
polymorphic nature of MHC molecules, the spectrum of peptides bound by these
molecules is highly variable. In Chapter 5 we will explore in greater detail
how peptide interacts with the β‐pleated sheet floor of MHC molecules, as these
interactions dramatically influence the type of peptides that can be presented
by particular molecules. The ongoing drive towards creating new MHC molecules, with
slightly altered peptide‐binding grooves, is akin to a genetic arms
race where the
immune system is constantly trying to keep one step ahead of
its foe. This genetic one‐upmanship has been termed pathogen‐driven
balancing selection because heterozygotes typically have a selective
advantage over homozygotes at a given locus.
The MHC
region represents an outstanding hotspot with mutation rates two orders of
magnitude higher than non‐ MHC loci. These multiple allelic forms can be
generated by a variety of mechanisms: point mutations, recombination,
homologous but unequal crossing over, and gene conversion.
The degree
of sequence homology and an increased occurrence of the dinucleotide motif
5′‐cytosine–guanine‐3′ (to produce what are referred to as CpG islands) seem to
be impor tant for gene conversion, and it has been suggested that this might
involve a DNA‐nicking activity that targets CpG‐rich DNA sequences. MHC genes
that lack these sequences, for example H‐2Ead and HLA‐DRA, do not
appear to undergo gene conversion, whereas those that possess CpG islands act
as either donors (e.g., H‐2Ebb, H‐2Q2k, H‐2Q10b),
acceptors (e.g., H‐2Ab) or both (e.g., H‐2Kk, HLA‐DQB1).
The large number of pseudogenes within the MHC may represent a stockpile of
genetic information for the generation of polymorphic diversity in the
“working” class I and class II molecules.
Nomenclature
As much of
the experimental work relating to the MHC is based on experiments in our little
laboratory friend, the mouse, it may be helpful to explain the nomenclature
used to describe the allelic genes and their products. If someone says to you
in an obscure language “we are having free elections,” you fail to understand,
not because the idea is complicated but because you do not comprehend the
language. It is much the same with the shorthand used to describe the H‐2
system, which looks unnecessarily frightening to the uninitiated. In order to
identify and compare allelic genes within the H‐2 complex in different strains,
it is usual to start with certain pure homozygous inbred strains, obtained by
successive brother–sister matings, to provide the prototypes. The collection of
genes in the H‐2 complex is called the haplotype and the
haplotype of each prototypic inbred strain will be allotted a given
superscript. For example, the DBA strain haplotype is designated H‐2d and
the genes constituting the complex are therefore H‐2Kd, H‐2Aad, H‐2Abd,
H‐2Dd, and so on; their products will be H‐2Kd, H‐2Ad, and H‐2Dd, and so
forth (Figure 4.27). When new strains are derived from these by genetic
recombination during breeding, they are assigned new haplotypes, but the
individual genes are designated by the haplotype of the prototype strain from
which they were derived. Thus the A/J strain produced by genetic cross‐over
during interbreeding between (H‐2k × H‐2d) F1 mice (Figure 4.28)
is arbitrarily assigned the haplotype H‐2a, but Table 4.4 shows that
individual genes in the complex are identified by the haplotype symbol of the original
parents.
Figure 4.27 How the
definition of H‐2 haplotype works. Pure strain mice homozygous for the
whole H‐2 region through prolonged brother–sister mating for at least 20
generations are each arbitrarily assigned a haplotype designated by a
superscript. Thus the particular set of alleles that happens to occur in the
strain named C57BL is assigned the haplotype H‐2b and the particular
nucleotide sequence of each allele in its MHC is labeled as geneb (e.g.,
H‐2Kb). It is obviously more convenient to describe a given allele by
the haplotype than to trot out its whole nucleotide sequence, and it is easier
to follow the reactions of cells of known H‐2 make‐up by using the
haplotype terminology (see, for example, the interpretation of the experiment
in Figure 4.28).
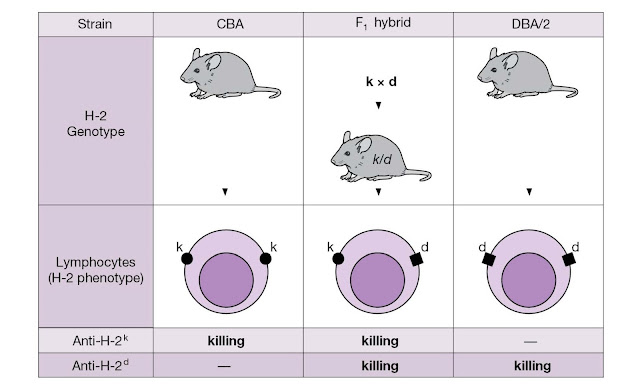
Figure 4.28 Inheritance and co‐dominant
expression of MHC genes. Each homozygous (pure) parental strain animal has two
identical chromosomes bearing the H‐2 haplotype, one paternal and the
other maternal. Thus in the present example we designate a strain that is H‐2k
as k/k. The first familial generation (F1) obtained by
crossing the pure parental strains CBA (H‐2k) and DBA/2 (H‐2d)
has the H‐2 genotype k/d. As 100% of F1 lymphocytes are killed in
the presence of complement by antibodies to H‐2k or to H‐2d (raised by
injecting H‐2k lymphocytes into an H‐2d animal and vice versa), the MHC
molecules encoded by both parental genes must be expressed on every lymphocyte.
The same holds true for other tissues in the body.
Pure strain
mice derived by prolonged brother sister mating are homozygous for each pair of
homologous chromosomes. Thus, in the present context, the haplotype of the MHC
derived from the mother will be identical to that from the father; animals of
the C57BL strain, for example, will each bear two chromosomes with the H‐2b haplotype
(see Table 4.4).
Let us see
how the MHC behaves when we cross two pure strains of haplotypes H‐2k and
H‐2d, respectively. We find that the lymphocytes of the offspring (the
F1 generation) all display both H‐2k and H‐2d molecules on their surface
(i.e., there is co‐dominant expression) (Figure 4.28). If we go
further and breed F1s together, the progeny have the genotypes k, k/d,
and d in the proportions to be expected if the haplotype
segregates as a single mendelian trait. This happens because the H‐2
complex spans 0.5 centimorgans, equivalent to a recombina tion frequency
between the K and D ends of 0.5%, and the haplotype tends to be
inherited en bloc. Only the relatively infrequent recombinations caused
by meiotic cross‐over events, as described for the A/J strain above, reveal the
complexity of the system.
The tissue distribution of MHC
molecules
Essentially,
all nucleated cells carry classical class I molecules. These are abundantly
expressed on both lymphoid and myeloid cells, less so on liver, lung, and
kidney and only sparsely on brain and skeletal muscle. In the human, the
surface of the placental extravillous cytotrophoblast lacks HLA‐A and ‐B,
although there is now some evidence that it may express HLA‐C. What is well
established is that the extravillous cytotrophoblast and other placental
tissues bear HLA‐G, a molecule that generally lacks allodeterminants and that
does not appear on most other body cells, except for medullary and sub
capsular epithelium in the thymus, and on blood monocytes following activation
with interferon‐γ. The role of HLA‐G in the placenta is not fully resolved, but
it appears to function as a replacement for classical class I molecules serving
to inhibit immune responses against paternal MHC alleles carried by the fetus.
Class II molecules, on the other hand, are highly restricted in their
expression, being present only on B‐cells, dendritic cells, macrophages, and
thymic epithelium. However, when activated by agents such as interferon‐γ,
capillary endothelia and many epithelial cells in tissues other than the thymus
express surface class II and increased levels of class I.
The nonclassical MHC and class I
chain‐related molecules
These
molecules include the CD1 family that utilize β2‐ microglobulin
and have an overall structure similar to the classical class I molecules
(Figure 4.29). They are, however, encoded by a set of genes on a different
chromosome to the MHC, namely on chromosome 1 in humans and chromosome 3 in the
mouse. Like its true MHC counterparts, CD1 is involved in the presentation of
antigens to T‐cells, but the anti gen‐binding groove is to some extent covered
over, contains mainly hydrophobic amino acids, and is accessible only through a
narrow entrance. Instead of binding peptide antigens, the CD1 molecules
generally present lipids or glycolipids. At least four different CD1 molecules
are found expressed on human cells; CD1a, b, and c are present on cortical thymocytes,
dendritic cells and a subset of B‐cells, whereas CD1d is expressed on
intestinal epithelium, hepatocytes, and all lymphoid and myeloid cells. Mice
appear to only express two different CD1 molecules that are both similar to the
human CD1d in structure and tissue distribution and are referred to as CD1d1
and CD1d2 (or CD1.1 and CD1.2).
Genes in the
MHC itself that encode nonclassical MHC molecules include the H‐2 T, H‐2Q,
and H‐2 M loci in mice, each of which encodes a number of different
molecules. The T22 and T10 molecules, for example, are induced by cellular
activation and are recognized directly by γδ TCR without a requirement for
antigen, possibly suggesting that they are involved in triggering
immunoregulatory γδ T‐cells. Other nonclassical class I molecules do bind
peptides, such as H‐2 M3 that presents N‐formylated peptides produced
either in mito chondria or by bacteria.
In the
human, HLA‐E binds a nine‐amino‐acid peptide derived from the
signal sequence of HLA‐A, ‐B, ‐C, and ‐G molecules, and is recognized by the
CD94/NKG2 receptors on NK cells and cytotoxic T‐cells, as well as by the αβ TCR
on some cytotoxic T‐cells. HLA‐E is upregulated when other HLA alleles
provide the appropriate leader peptides, thereby allowing NK cells to monitor
the expression of polymorphic class I molecules using a single receptor. The
murine homolog, Qa‐1, has a similar function.
The
stress‐inducible MICA and MICB (MHC class I chain‐related molecules) have the
same domain structure as classical class I and display a relatively high level
of polymorphism. They are present on epithelial cells, mainly in the
gastrointestinal tract and in the thymic cortex, and are recog nized by the
NKG2D‐activating molecule. One possible role for this interaction is in the
promotion of NK cell and T‐cell antitumor responses.
The function
of HLA‐F is unclear, although its expression in placental
trophoblasts has led some to suggest that it may play a role in protecting the
developing fetus from attack by the maternal immune system. A more definitive
role for HLA‐G in this context has been found. This HLA molecule
is also preferentially expressed on placental trophoblast cells where it plays
a role in shielding the fetus from the unwanted attentions of the maternal NK
cells and cytotoxic T‐cells. It has long been a puzzle why mothers tolerate
their genetically non‐identical fetuses, as one would normally expect a strong
immune response to foreign (i.e., paternal) HLA molecules. Although this is
partially solved through downregulation of the expression of MHC class I A, B,
and C molecules on placenta, this would normally attract the attentions of NK
cells on the prowl for cells with such missing‐self characteristics, as we
discussed earlier when dealing with NK receptors. HLA‐G expression on the
placen tal–maternal trophoblast interface appears to be a solution to this.
The interaction between the immunoglobulin‐like tran script‐2 (ILT2) molecule
on NK cells, which is an inhibitory NK receptor, with HLA‐G expressed on
placental trophoblasts confers protection against NK cell‐mediated cytolysis.
HFE, previously referred to
as HLA‐H, possesses an extremely narrow groove that is unable to bind peptides,
and it may serve no role in immune defense. However, it binds to the transferrin
receptor and appears to be involved in iron uptake. A point mutation (C282Y) in
HFE is found in 70–90% of patients with hereditary hemochromatosis.
Figure 4.29 Comparison of the crystal
structures of CD1 and MHC class I. (a) Backbone ribbon diagram of mouse CD1d1
(red, α‐helices; blue, β‐strands). (b) Ribbon diagram of the mouse MHC class I
molecule H‐2Kb (cyan, α‐helices; green, β‐strands). (c) Superposition using
alignment of β2‐microglobulin highlights some of the differences between CD1d1
and H‐2Kb. Note in particular the shifting of the α‐helices. This produces a
deeper and more voluminous groove in CD1d1, which is narrower at its entrance
compared with H‐2Kb. (Source: Porcelli ) Immunology Today 19,
362. Reproduced with permission of Elsevier.)
Nonclassical MHC molecules may be
the precursors to classical MHC molecules
Analysis of
vertebrate genomes suggests that invariant nonclas sical MHC molecules are
probably the primordial forerunner to modern polymorphic MHC class I and class
II molecules and rather than playing a role in antigen presentation, these
molecules were most likely used as primitive “danger signals” involved in
conveying stress signals to innate immune cells. Thus, expression of these
molecules on the cell surface signified a stressed or potentially transformed
cell that should be eliminated in the interests of overall organismal fitness.
During the course of evolution, such molecules then most likely evolved the
ability to bind self peptides, which were initially relatively invariant,
followed by the ability to bind highly variable peptides, as we now see with
classical MHC class I and class II gene products. The appearance of polymorphic
MHC molecules, as a consequence of gene duplication events followed by
divergence, would have enabled much greater diversity in the range of peptides
bound by these molecules. Thus, invariant MHC‐like molecules (such as HLA‐E,
‐F, ‐G, and MICA, MICB) tend not to have antigen‐presenting functions, but
perform homeostatic or regulatory roles, permitting cells of the innate immune
system to monitor cell health in a relatively antigen‐nonspecific way.
A good
example, which was discussed in the context of NK receptors but is worth going
over again, is the HLA‐E molecule that binds a nine‐amino‐acid peptide derived
from the signal sequence of HLA‐A, ‐B, and ‐C molecules. Should HLA‐E– peptide
complexes be absent from cells, this suggests that an infectious agent may be
present or that cells are stressed in some way. This results in activation of
NK cells via the activat ing CD94/NKG2 receptors, with consequent NK‐mediated
killing of such cells. In the absence of class I leader peptides, HLA‐E can be
stabilized on the surface of stressed cells by heat‐shock treatment because the
HSP‐60 signal peptide can also bind in place of HLA class I peptides. However,
such HLA‐E/ HSP‐60 leader peptide complexes fail to be recognized by the
CD94/NKG2 receptor, once again precipitating attack by the NK cell. Thus, cell
stress can override the presentation of class I‐derived peptides through
competition for HSP‐60‐derived peptides that would not normally be present at
levels high enough to compete effectively in unstressed cells. If this isn’t a clever
molecular security system, we don’t know what is.