The T‐Cell Surface Receptor For Antigen (TCR)
As alluded
to earlier, T‐cells interact with antigen in a manner that is quite distinct
from the way in which B‐cells do; the receptors that most T‐cells are equipped
with cannot directly engage soluble antigens but instead “see” fragments of
antigen that are immobilized within a narrow groove on the surface of MHC
molecules (Figure 4.1b).
As we shall discuss in detail in Chapter 5, MHC
molecules bind to short 8–20 amino acid long peptide fragments that represent
“quality control” samples of the proteins a cell is expressing at any given
time, or what it has internalized through phagocytosis, depending on the type
of MHC molecule. In this way, T‐cells can effectively inspect what is going on,
antigenically speaking, within a cell at any given moment by surveying the
range of peptides being presented within MHC molecules. Another major
difference between B‐ and T‐cell receptors is that T‐cells cannot secrete their
receptor molecules in the way that B‐cells can switch production of Ig from a
membrane‐bound form to a secreted form. These differences aside, T‐cell
receptors are structurally quite similar to antibody as they are also
built from modules that are based upon the immunoglobulin fold.
Before we
explore the structural aspects of T‐cell receptors, please keep in mind that
the practical function of these recep tors is to enable a T‐cell to probe the
surfaces of cells looking for nonself peptides. If a T‐cell finds a peptide–MHC
combination that is a good match for its TCR it will become acti vated,
undergo clonal expansion, and differentiate to a mature effector T‐cell capable
of joining the fight against the infectious agent generating these nonself
peptides. In practice, such an eventuality is a very low probability event
because, as we shall see, TCRs are generated in such a way as to produce an
enor mous variety of these receptors, each with their own exquisite
specificity for a particular peptide–MHC combination. Moreover, because the
majority of peptides presented on MHC molecules at any one time will be derived
from self (unless the antigen‐presenting cell is infected with a
microorganism), this further reduces the probability of a T‐cell encountering a
per fect nonself peptide–MHC combination to trigger a response.
Milestone 4.1 The T‐cell receptor
As T‐lymphocytes respond by activation and
proliferation when they contact antigen presented by cells such as macrophages,
it seemed reasonable to postulate that they do so by receptors on their
surface. In any case, it would be difficult to fit T‐cells into the clonal
selection club if they lacked such receptors. Guided by Occam’s razor (the law
of parsimony, which contends that it is the aim of science to present the facts
of nature in the simplest and most economical conceptual formulations), most
investigators plumped for the hypothesis that nature would not indulge in the
extravagance of evolving two utterly separate molecular recognition species for
B‐ and T‐cells, and many fruitless years were spent looking for the “Holy
Grail” of the T‐cell receptor with anti‐immunoglobulin serums or monoclonal
antibodies. Success only came when a monoclonal antibody directed to the
idiotype of a T‐cell was used to block the response to antigen. This was
identified by its ability to block one individual T‐cell clone out of a large
number, and it was correctly assumed that the structure permitting this
selectivity would be the combining site for antigen on the T‐cell receptor. Immunoprecipitation
with this antibody brought down a disulfide‐linked heterodimer composed of
40–44 kDa subunits (Figure M4.1.1).
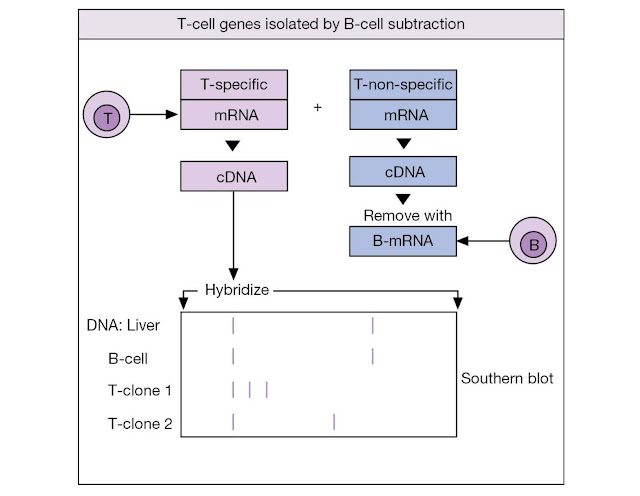
The other approach went directly for the genes,
arguing as follows. The T‐cell receptor should be an integral membrane protein
not present in B‐cells. Hence, T‐cell polysomal mRNA from the endoplasmic
reticulum, which should provide an abundant source of the appropriate
transcript, was used to prepare cDNA from which genes common to B‐ and T‐cells
were subtracted by hybridization to B‐cell mRNA. The resulting T‐specific
clones were used to probe for a T‐cell gene that is rearranged in all
functionally mature T‐cells but is in its germline configuration in all other
cell types (Figure M4.1.2). In such a way were the genes encoding the β‐subunit
of the T‐cell receptor uncovered.
The receptor for antigen is a
transmembrane heterodimer
Identification
of the TCR proved more difficult than initially anticipated (Milestone 4.1),
but eventually the receptor was found to be a membrane‐bound molecule composed
of two disulfide‐linked chains, α and β. Each chain folds into two Ig‐ like
domains, one having a relatively invariant structure and the other exhibiting a
high degree of variability, so that the αβ TCR has a structure really quite
closely resembling an Ig Fab fragment. This analogy stretches even further –
each of the two variable regions has three hypervariable regions (or
complementarity determining regions, CDRs) that X‐ray diffraction data have
defined as incorporating the amino acids that make contact with the peptide–MHC
ligand. Plasticity of the CDR loops is an important factor, enabling TCRs to
mold around structurally diverse peptide MHC combinations.
Although the
manner in which the TCR makes contact with peptide–MHC is still not fully
understood, it appears that in some TCRs CDRs 1 and 2 of the TCR bear much of
the responsibility for making contact with the MHC molecule itself, while CDR3
makes contact with the peptide; however in other TCRs the
reverse is true. Whatever CDRs bear
the responsibility for contacting MHC versus peptide, it is clear that
these are the recognition components of the receptor and so it follows that it
is here that much of the variability is seen between TCRs, as we shall discuss
later.
Both α and β
chains are required for antigen specificity as shown by transfection of the
T‐receptor genes from a cytotoxic T‐cell clone specific for fluorescein to
another clone of a different specificity; when it expressed the new α and β
genes, the transfected clone acquired the ability to lyse the fluoresceinated
target cells. Another type of experiment utilized T‐cell hybridomas formed by
fusing single antigen‐specific T‐cells with T‐cell tumors to achieve
“immortality.” One hybridoma recog nizing chicken ovalbumin, presented by a
macrophage, gave rise spontaneously to two variants, one of which lost the
chromosome encoding the α chain, and the other, the β chain. Neither variant
recognized antigen but, when they were physically fused together, each supplied
the complem ntary receptor chain, and reactivity with antigen was restored.
CD4 and CD8 molecules act as
co‐receptors for TCRs
In addition
to the TCR, the majority of peripheral T‐cells also express one or other of the
membrane proteins CD4 or CD8 that act as co‐receptors
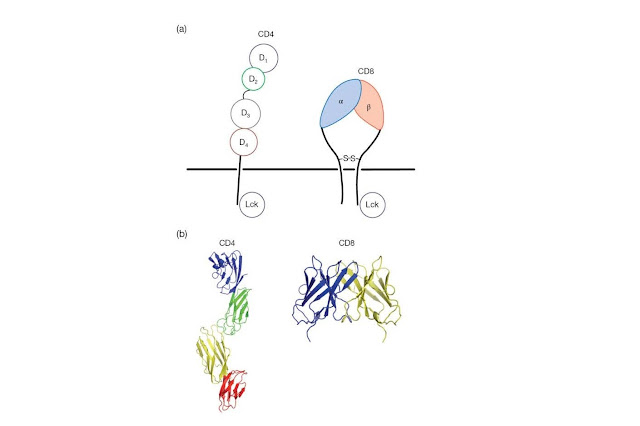
for MHC molecules (Figure 4.7). CD4 is a single‐chain polypeptide containing
four Ig‐like domains packed tightly together to form an extended rod that
projects from the T‐cell surface. The cytoplasmic tail of the CD4 molecule is
important for TCR signaling as this region is constitutively bound by a protein
tyrosine kinase, Lck, that initiates the signal transduction
cascade that follows upon encounter of a T‐cell with antigen (Figure 4.8). CD8
plays a similar role to CD4, as it also binds Lck and recruits this kinase to
the TCR complex, but is structurally quite distinct; CD8 is a disulfidelinked
heterodimer of α and β chains, each of which contains a single Ig‐like domain
connected to an extended and heavily glycosylated polypeptide projecting from
the T‐cell surface (Figure 4.7).
CD4 and CD8
molecules play important roles in antigen recognition by T‐cells as these
molecules dictate whether a T‐ cell can recognize antigen presented by MHC
molecules that obtain their peptide antigens primarily from intracellular (MHC
class I), or extracellular (MHC class II), sources. This
has major functional implications for the T‐cell, as those lymphocytes that
become activated upon encounter with antigen presented within MHC class I
molecules (CD8+ T‐cells) invariably become cytotoxic T‐cells, and
those that are activated by peptides presented by MHC class II molecules (CD4+
T‐cells) become helper T‐cells (see Figure 7.1).
There are two classes of T‐cell
receptors
Not long
after the breakthrough in identifying the αβ TCR, reports came of the existence
of a second type of receptor composed of γ and δ chains. As it appears earlier
in thymic ontogeny, the γδ receptor is sometimes referred to as TCR1 and
the αβ receptor as TCR2.
The γδ cells
make up only 1–5% of the T‐cells that circulate in blood and peripheral organs
of most adult animals; however these cells are much more common in
epithelial‐rich tissues such as the skin, intestine, reproductive tract, and
the lungs where they can comprise almost 50% of the T‐cell population. It
cannot be denied that γδ T‐cells are somewhat of an oddity among T‐cells;
unlike αβ T‐cells, γδ cells do not appear to require antigen to be presented
within the context of MHC molecules and are thought to be able to recognize
soluble antigen akin to B‐cells. Perhaps because of this lack of dependence on
MHC for antigen presentation, the majority of γδ T‐cells do not express either
of the MHC co‐receptors, CD4 or CD8 (Table 4.1).
The
mechanism of antigen recognition by γδ T‐cells is still somewhat mysterious but
these cells are known to be able to interact with MHC‐related molecules, such
as the mouse T10 and T22 proteins, in a manner that does not require antigen.
Because the latter MHC‐like molecules are upregulated upon may have an
important immunoregulatory function; by becoming activated by molecules that
appear on activated T‐ cells, γδ T‐cells may help to regulate immune responses
in a positive or negative
manner. γδ T‐cells can also recognize pathogen‐derived lipids, organic
phosphoesters, nucleotide conjugates, and other nonpeptide ligands.
Certain γδ
T‐cells (the Vγ1 Vδ1 subset, which are enriched in epithelial tissues) also
share some of the same recognition features of NK cells of the innate immune
system, as they can both recognize the MHC class I‐like proteins MICA and MICB,
which do not function as antigen‐presenting molecules. Rather, MICA and MICB
are typically present at low levels on epithelial tissues but are upregulated in
response to cellular stress, including heat shock and DNA damage. Infection
with cytomegalovirus or Mycobacterium tuberculosis is also capable of
inducing the surface appearance of these primitive MHC‐like molecules and other
stress‐inducible γδ T‐cell ligands are almost certain to exist. As we shall see
later in this chapter, MICA and MICB are also used by NK cells as activation
ligands, although in this case a very different receptor is responsible.
The encoding of TCRs is similar to
that of immunoglobulins
The gene
segments encoding the TCR β chains follow a broadly similar arrangement of V,
D, J, and constant segments to that described for the
immunoglobulins (Figure 4.9). In a parallel fashion, as an immunocompetent
T‐cell is formed, rearrangement of V, D, and J genes
occurs to form a continuous VDJ sequence. The firmest evidence that B‐
and T‐cells use similar recombination mechanisms comes from mice with severe
combined immunodeficiency (SCID) that have a single autosomal recessive defect
preventing successful recombination of V, D, and J segments.
Homozygous mutants fail to develop immuno competent B‐ and T‐cells and
identical sequence defects in VDJ joint formation are seen in both
pre‐B‐ and pre‐T‐cell lines.
Looking
first at the β chain cluster, one of the two Dβ genes rearranges next to
one of the Jβ genes. Note that, because of the way the genes are
organized, the first Dβ gene, Dβ1, can utilize any of the 13 Jβ
genes, but Dβ2 can only choose from the seven Jβ2 genes
(Figure 4.9). Next, one of the 50 or so Vβ genes is rear ranged to the
preformed DβJβ segment. Variability in junction formation and
the random insertion of nucleotides to create N‐region diversity
either side of the D segment mirror the same phenomenon seen with Ig
gene rearrangements. Sequence analysis emphasizes the analogy with the antibody
molecule; each V segment contains two hypervariable regions, while the DJ
junctional sequence provides the very hypervariable CDR3
structure, making a total of six potential CDRs for antigen bind ing in each
TCR (Figure 4.10). As in the synthesis of antibody, the intron between VDJ and
C is spliced out of the mRNA before translation with the restriction that
rearrangements involving genes in the Dβ2Jβ2 cluster can only link to Cβ2.
All the
other chains of the TCRs are encoded by genes formed through similar
translocations. The α chain gene pool lacks D segments but
possesses a prodigious number of J segments. The number of Vγ and
Vδ genes is small in comparison with Vα and Vβ. Like the α
chain pool, the β chain cluster has no D segments. The
awkward location of the δ locus embedded within the α gene
cluster results in T‐cells that have undergone Vα–Jα combination having
no δ genes on the rearranged chromosome; in other words, the δ genes
are completely excised.
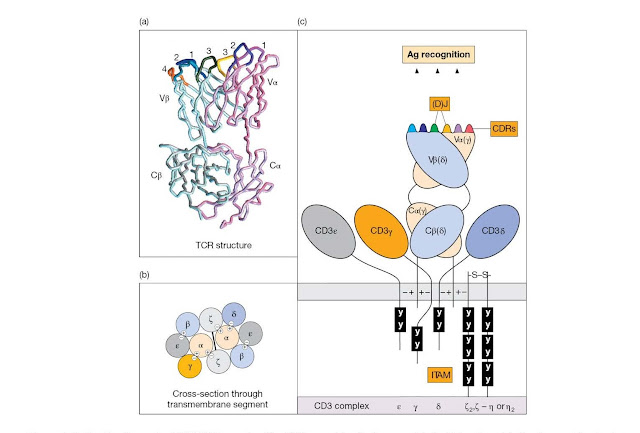
Figure 4.10 The
T‐cell receptor (TCR)/CD3 complex. The TCR resembles the immunoglobulin Fab
antigen‐binding fragment in structure. The variable and constant segments of
the TCR α and β chains (VαCα/VβCβ), and of the corresponding γ and δ chains of
the γδ TCR, belong structurally to the immunoglobulin‐type domain family. (a)
In the model the α chain CDRs are colored magenta (CDR1), purple (CDR2), and
yellow (CDR3), whilst the β chain CDRs are cyan (CDR1), navy blue (CDR2) and
green (CDR3). The fourth hypervariable region of the β chain (CDR4), which
constitutes part of the binding site for some superantigens, is colored orange.
(Reproduced from Garcia K. et al. (1998) Science 279, 1166; with
permission.) The TCR α and β CDR3 loops encoded by (D)J genes are both
short; the TCR γ CDR3 is also short with a narrow length distribution, but the δ
loop is long with a broad length distribution, resembling the Ig light and
heavy chain CDR3s, respectively. (b) The TCRs may be expressed in pairs linked
to the CD3 complex. Negative charges on trans membrane segments of the
invariant chains of the CD3 complex contact the opposite charges on the TCR Cα and
Cβ chains conceivably as depicted. (c) The cytoplasmic domains of the CD3
peptide chains contain immunoreceptor tyrosine‐based activation motifs (ITAMs;
see BCR, Figure 4.4) that contact src protein tyrosine kinases. Try not to
confuse the TCR γδ and the CD3 γδ chains.
The CD3 complex is an integral part
of the T‐cell receptor
The T‐cell
antigen recognition complex and its B‐cell counterpart can be likened to army
scouts whose job is to let the main battalion know when the enemy has been
sighted. When the TCR “sights the enemy” (i.e., ligates antigen), it relays a
signal through an associated complex of transmembrane polypeptides (CD3)
to the interior of the T‐lymphocyte, instructing it to awaken from its
slumbering G0 state and do something useful – like becoming an effector cell.
In all immunocompetent T‐cells, the TCR is noncovalently but still intimately
linked with CD3 in a complex that, as current wisdom has it, may contain two
heterodimeric TCR αβ or γδ recognition units closely apposed to one molecule of
the invariant CD3 polypep tide chains γ and δ, two molecules of CD3ε, plus the
disulfidelinked ζ–ζ dimer. The total complex therefore has the structure
TCR2–CD3γδε2–ζ2 (Figure 4.8 and Figure 4.10b).
Similar to
the BCR‐associated Ig–α/β heterodimer, the CD3 chains also contain one or more
ITAMs and these motifs, once again, are instrumental in the propagation of
activation signals into the lymphocyte. Upon encounter of the TCR with
peptide–MHC, the ITAMs within the CD3 complex become phosphorylated at tyrosine
residues; these then act as a platform for the recruitment of a veritable
multitude of phosphotyrosine‐binding proteins that further disseminate the
signal throughout the T‐cell. It is here that the role of the CD4 and CD8
co‐receptors becomes apparent; phosphorylation of the ITAMs within the CD3 ζ
(zeta) chain is accomplished by the Lck tyrosine kinase that, you may recall,
is associated with the cytoplasmic tails of CD4 and CD8 (Figure 4.7 and Figure
4.8). In mice, either or both of the ζ chains can be replaced by a splice variant
from the ζ gene termed η. The ζ chain also associates with the FcγRIIIA
receptor in natural killer (NK) cells where it functions as part of the signal
transduction mechanism in that context also. We shall discuss TCR‐initiated
signal transduction in much greater detail in Chapter 7.