The Generation Of
Diversity For Antigen Recognition
We know that
the immune system has to be capable of recognizing virtually any pathogen that
has arisen or might arise. The awesome genetic solution to this problem of anticipating
an unpredictable future involves the generation of millions of different
specific antigen receptors, probably vastly more than the lifetime needs of the
individual. As this greatly exceeds the estimated number of 25 000–30 000 genes
in the human body, there are some clever ways to generate all this diversity,
particularly as the total number of V, D, J, and C genes
in an individual human coding for antibodies and TCRs is only around 400. Let’s
revisit the genetics of antibody diversity, and explore the enormous
similarities, and occasional differences, seen with the mechanisms employed to
generate TCR diversity.
Intrachain amplification of
diversity
Random VDJ combination increases
diversity geometrically
We saw in
Chapter 3 that, just as we can use a relatively small number of different
building units in a child’s construction set such as LEGO® to create a rich
variety of architectural masterpieces, so the individual receptor gene segments
can be viewed as building blocks to fashion a multiplicity of antigen specific
receptors for both B‐ and T‐cells. The immunoglobulin light chain variable
regions are created from V and J segments, and the heavy chain
variable regions from V, D, and J segments. Likewise, for
both the αβ and γδ T‐cell receptors the variable region of one of the chains (α
or γ) is encoded by a V and a J segment, whereas the variable
region of the other chain (β or δ) is additionally encoded by a D segment.
As for immunoglobulin genes, the enzymes RAG‐1 and RAG‐2 recognize recombination
signal sequences (RSSs) adjacent to the coding sequences of the TCR V, D,
and J gene segments. The RSSs again consist of conserved heptamers and
nonamers separated by spacers of either 12 or 23 base‐pairs and are found at
the 3′ side of each V segment, on both the 5′ and 3′ sides of each D segment,
and at the 5′ of each J segment. Incorporation of a D segment is
always included in the rearrangement; Vβ cannot join directly to Jβ, nor Vδ
directly to Jδ. To see how sequence diversity is generated for TCR, let us take
the αβ TCR as an example (Table 4.2). Although the precise number of gene
segments varies from one individual to another, there are typically around 75 Vα
gene segments and 60 Jα gene segments. If there were entirely random
joining of any one V to any one J segment, we would have
the possibility of generating 4500 VJ combinations (75 × 60). Regarding
the TCR β‐chain, there are approximately 50 Vβ genes that lie upstream
of two clusters of DβJβ genes, each of which is associated with a Cβ gene (Figure
4.11). The first cluster, associated with Cβ1, has a single Dβ1 gene and 6 Jβ1
genes, whereas the second cluster associated with Cβ2 again has a single Dβ
gene (Dβ2) with 7 Jβ2.
The Dβ1
segment can combine with any of the 50 Vβ genes and with any of the 13 Jβ1
and Jβ2 genes (Figure 4.11). β2 behaves similarly but can only
combine with one of the 7 downstream Jβ2 genes. This provides
1000 different possible VDJ combinations for the TCR β‐chain. Therefore,
although the TCR α and β chain V, D, and J genes add up
arithmetically to just 200, they produce a vast number of different α and β
variable regions by geometric recombination of the basic
elements. But, as with immunoglobulin gene rearrangement, that is only the
beginning.
Figure
4.11 Rearrangement of the T‐cell receptor β‐chain gene locus. In this example
Dβ1 has rearranged to Jβ2.2, and then the Vβ2 gene selected out of the 50 or so
(Vβn) Vβ genes. If the same V and D segments had been used, but this time Jβ1.4
had been employed, then the Cβ1 gene segment would have been utilized instead
of Cβ2.
Playing with the junctions
Another ploy
to squeeze more variation out of the germline repertoire that is used by both
the TCR and the immunoglobulin genes (see Figure 3.25) involves variable
boundary recombinations of V, D, and J to produce
different junctional sequences (Figure 4.12.).
As discussed
in Chapter 3, further diversity results from the generation of palindromic
sequences (P‐elements) arising from the formation of hairpin structures during
the recombination process and from the insertion of nucleotides at the N region
between the V, D, and J segments, a process associated
with the expression of terminal deoxynucleotidyl transferase. While these
mechanisms add nucleotides to the sequence, yet more diversity can be created
by nucleases chewing away at the exposed strand ends to remove nucleotides.
These maneuvers again greatly increase the repertoire, especially important for
the TCR γ and δ genes, which are otherwise rather limited in number.
Additional
mechanisms relate specifically to the D‐region sequence: particularly in
the case of the TCR δ genes, where the D segment can be read in three
different reading frames and two D segments can join together. Such DD
combinations produce a longer third complementarity determining region
(CDR3) than is found in other TCR or antibody molecules.
As the CDR3
in the various receptor chains is essentially composed of the regions between
the V(D)J segments, where junctional diversity mechanisms can introduce
a very high degree of amino acid variability, one can see why it is that this hypervariable
loop usually contributes the most to determining the fine antigen‐binding
specificity of these molecules.
Figure
4.12 Junctional diversity between a TCR V α and J α germline segment producing
three variant protein sequences. The nucleotide triplet that is
spliced out is colored the darker blue. For TCR β chain and Ig heavy chain
genes junctional diversity can apply to V, D, and J segments.
Receptor editing
Recent
observations have established that lymphocytes are not necessarily stuck with
the antigen receptor they initially make: if they don’t like it they can change
it. The replacement of an undesired receptor with one that has more acceptable
characteristics is referred to as receptor editing. This process
has been described for both immunoglobulins and for TCR, allowing the
replacement of either nonfunctional rearrangements or autoreactive
specificities. Furthermore, receptor editing in the periphery may rescue
low‐affinity B‐cells from apoptotic cell death by replacing a low‐affinity
receptor with a selectable one of higher affinity. That this does indeed occur
in the periphery is strongly supported by the finding that mature B‐cells in
germinal centers can express RAG‐1 and RAG‐2 that mediate the rearrangement
process.
But how does
this receptor editing work? Well, in the case of the receptor chains that lack D
gene segments, namely the immunoglobulin light chain and the TCR α chain, a
secondary rearrangement may occur by a V gene segment upstream of the previously
rearranged VJ segment recombining to a 3′ J gene sequence, both
of these segments having intact RSSs that are compatible (Figure 4.13a).
However, for immunoglobulin heavy chains and TCR β deletes all of the D segment‐associated
RSSs (Figure 4.13b). Because VH and JH both have 23 basepair
spacers in their RSSs, they cannot recombine: that would break the 12/23 rule.
This apparent obstacle to receptor editing of these chains may be overcome by
the presence of a sequence near the 3′ end of the V coding sequences
that can function as a surrogate RSS, such that the new V segment would
simply replace the previously rearranged V, maintaining the same D and
J sequence (Figure 4.13b). This is probably a relatively inefficient
process and receptor editing may therefore occur more readily in immunoglobulin
light chains and TCR α chains than in
immunoglobulin heavy chains and TCR β chains. Indeed, it has been suggested
that the TCR α chain may undergo a series of rearrangements, continuously
deleting previously functionally rearranged VJ segments until a
selectable TCR is produced.
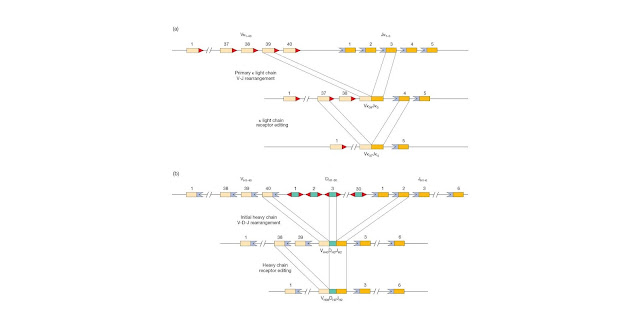
Figure 4.13 Receptor
editing. (a) For immunoglobulin light chain or TCR α chain the recombination
signal sequences (RSSs; heptamer– nonamer motifs) at the 3′ end of each
variable (V) segment and the 5′ of each joining (J) segment are
compatible with each other and therefore an entirely new rearrangement can
potentially occur as shown. This would result in a receptor with a different
light chain variable sequence (in this example Vκ37Jκ4 replacing Vκ39Jκ3)
together with the original heavy chain. (b) With respect to the immunoglobulin
heavy chain or TCR β chain the organization of the heptamer–nonamer sequences
in the RSS precludes a V segment directly recombining with the J segment.
This is the so‐called 12/23 rule whereby the heptamer–nonamer sequences
associated with a 23 base‐pair spacer (colored violet) can only base‐pair with
heptamer–nonamer sequences containing a 12 base‐pair spacer (colored red). The
heavy chain V and J both have an RSS with a 23 base‐pair spacer
and so this is a nonstarter. Furthermore, all the unrearranged D segments
have been deleted so that there are no 12 base‐pair spacers remaining. This
apparent bar to secondary rearrangement is probably overcome by the presence of
an RSS‐like sequence near the 3′ end of the V gene coding sequences, so
that only the V gene segment is replaced (in the example shown, the
sequence VH38DH3JH2 replaces VH40DH3JH2).
Recognition of the correct
genomic regions by the RAG recombinase
A question
that is only now being resolved is how the RAG‐1/ RAG‐2 recombinase selects the
correct genomic regions to target for recombination. Clearly it would be
disastrous were this complex able to access all DNA, randomly leaving
double‐stranded breaks in its wake. One mechanism of protection is to induce
RAG expression only where and when it is needed, but this does not explain how
the RAG complex is targeted only to Ig and TCR loci in the cells in which it is
expressed. This puzzle is explained by observations suggesting that alterations
to histones – the proteins upon which DNA is packaged – flag particular
loci for binding of the RAG complex. Recent studies have shown that histone H3
that has been modified by tri methylation on lysine at position 4 (H3K4me3)
acts as a bind ing site for RAG‐2. Thus, genomic regions that are poised for
VDJ recombination are located close to H3K4me3 histone “marks.” Consistent with
this idea, experimental ablation of H3K4me3 marks results in greatly impaired
V(D)J recombination. But the H3K4me3 mark is found at many more sites
throughout the genome than there are antigen receptor loci, so how does the
RAG‐1/RAG‐2 complex find the correct sites? The answer seems to be that the
specificity of RAG‐1 for RSS sites, combined with that of RAG‐2 for H3K4me3
chromatin marks, may act as a clamp that guides the recombinase to the right
locations. Binding of the RAG complex to the H3K4me3 mark may also activate the
recombinase activity of RAG‐1 through an allosteric mechanism, increasing the
catalytic activ ity of the complex when it has been positioned at the correct
location.
Interchain amplification
The immune
system took an ingenious step forward when two different types of chain were
utilized for the recognition molecules because the combination produces not
only a larger combining site with potentially greater affinity, but also new
variability. Heavy–light chain pairing among immunoglobulins appears to be
largely random and therefore two B‐cells can employ the same heavy chain but
different light chains. This route to producing antibodies of differing
specificity is easily seen in vitro where shuffling different
recombinant light chains against the same heavy chain can be used to either
fine‐tune, or sometimes even alter, the specificity of the final antibody. In
general, the available evidence suggests that in vivo the major
contribution to diversity and specificity comes from the heavy chain, perhaps
not unrelated to the fact that the heavy chain CDR3 gets off to a head start in
the race for diversity being, as it is, encoded by the junctions between three
gene segments: V, D, and J.
This random
association between TCR γ and δ chains, TCR α and β chains, and Ig heavy and
light chains yields a further geometric increase in diversity. From Table 4.2
it can be seen that approximately 230 functional TCR and 153 functional Ig
germline segments can give rise to 4.5 million and 2.3 million different
combinations, respectively, by straightforward associations without taking
into account all of the fancy junctional mechanisms described above. Hats off
to evolution!
Somatic hypermutation
As discussed
in Chapter 3, there is inescapable evidence that immunoglobulin V‐region
genes can undergo significant somatic hypermutation. Analysis of
18 murine λ myelomas revealed 12 with identical structure, four showing just
one amino acid change, one with two changes and one with four changes, all
within the hypervariable regions and indicative of somatic hypermutation of the
single mouse λ germline gene. In another study, following immunization with
pneumococcal antigen, a single germline T15 VH gene gave rise by
mutation to several different VH genes all encoding phosphorylcholine
antibodies (Figure 4.14).
A number of
features of this somatic diversification phe nomenon are worth revisiting. The
mutations are the result of single nucleotide substitutions, they are
restricted to the variable as distinct from the constant region and occur in
both framework and hypervariable regions. The mutation rate is remarkably high,
approximately 1 × 10−3 per base‐pair per generation, which is approximately a
million times higher than for other mammalian genes. In addition, the
mutational mechanism is bound up in some way with class switch recombination as
the enzyme activation‐induced cytidine deaminase (AID)
is required for both processes and hypermutation is more frequent in IgG and
IgA than in IgM antibodies, affecting both heavy (Figure 4.14) and light
chains. However, VH genes are, on average more mutated than VL
genes. This might be a consequence of receptor editing acting more
frequently on light chains, as this would have the effect of wiping the slate
clean with respect to light chain V gene mutations while maintaining
already accumulated heavy chain V gene point mutations.
As we
outlined in Chapter 3, AID initiates both class switch recombination as well as
somatic hypermutation through deaminating deoxycytidine within certain DNA
hotspots that are characterized by the presence of WRC sequences (W = A or T, R
= purine, and C is the deoxycytidine that becomes deaminated). Although the
target of AID was initially thought to be RNA, more recent evidence suggests
that this enzyme works directly on DNA, although RNA editing is not ruled out.
Deamination of deoxycytidine changes this base to a deoxyuracil that would
normally be repaired by mismatch repair enzymes but, for reasons that are not
yet fully understood, can result in removal of the mismatched uracil that
generates a gap that is filled in by an errorprone polymerase to generate a
point mutation at this position and can also mutate surrounding bases. It
remains unclear how AID is targeted to the correct locations within V regions
of rearranged Ig genes, to ensure that mutations are not inadvertently
introduced at other loci, but similar to the RAG recombinase, this might
involve specific histone modifications. Hyperacetylated versions of histones H3
and H4 appear to be more abundant in mutating V regions than in the C regions
of Ig genes. This observation, coupled with observations that AID is recruited
to actively transcribing Ig genes by proteins that bind to CAGGTG sequences
found in all Ig transcriptional enhancers, suggests a possible mechanism. Thus,
the combination of the CAGGTG sequence motif, coupled with the modified
histones discussed above, may position AID at the correct locations from which
to operate.
Somatic
hypermutation does not appear to add significantly to the repertoire available
in the early phases of the primary response, but occurs during the generation
of memory and is responsible for tuning the response towards higher affinity.
Recently,
data have been put forward suggesting that there is yet another mechanism for
creating further diversity. This involves the insertion or deletion of short
stretches of nucleotides within the immunoglobulin V gene sequence of
both heavy and light chains. This mechanism would have an intermediate effect
on antigen recognition, being more dramatic than single point mutation, but
considerably more subtle than receptor editing. In one study, a reverse
transcriptase‐polymerase chain reaction (RT‐PCR) was employed to amplify the
expressed VH and VL genes from 365 IgG+
B‐cells and it was shown that 6.5% of the cells contained nucleotide insertions
or deletions. The transcripts were left in‐frame and no stop codons were
introduced by these modifications. The percentage of cells containing these
alterations is likely to be an underestimate. All the insertions and deletions
were in, or near to, CDR1 and/or CDR2. N‐region diversity of the CDR3 meant
that it was not possible to analyze the third hypervariable region for
insertions/deletions of this type and therefore these would be missed in the
analysis. The fact that the alterations were associated with CDRs does suggest
that the B‐cells had been subjected to selection by antigen. It was also
notable that the insertions/deletions occurred at known hotspots for somatic
point mutation, and the same error‐prone DNA polymerase responsible for somatic
hypermutation may also be involved here. The sequences were often a duplication
of an adjacent sequence in the case of insertions or a deletion of a known
repeated sequence. This type of modification may, like receptor editing, play a
major role in eliminating autoreactivity and also in enhancing antibody
affinity.
T‐cell receptor genes, on the other hand, do
not generally undergo somatic hypermutation. It has been argued that
this would be a useful safety measure as T‐cells are positively selected in the
thymus for weak reactions with self MHC, so that mutations could readily lead
to the emergence of high‐ affinity autoreactive receptors and autoimmunity.
One may ask
how it is that this array of germline genes is protected from genetic drift.
With a library of 390 or so functional V, D, and J genes,
selection would act only weakly on any single gene that had been functionally
crippled by mutation and this implies that a major part of the library could be
lost before evolutionary forces operated. One idea is that each subfamily of
related V genes contains a prototype coding for an antibody
indispensable for protection against some common pathogen, so that mutation in
this gene would put the host at a disadvantage and would therefore be selected
against. If any of the other closely related genes in its set became defective
through mutation, this indispensable gene could repair them by gene conversion,
a mechanism in which two genes interact in such a way that the nucleotide
sequence of part or all of one becomes identical to that of the other. Although
gene conver sion has been invoked to account for the diversification of MHC
genes, it can also act on other families of genes to main tain a degree of
sequence homogeneity. Certainly it is used extensively by, for example,
chickens and rabbits, in order to generate immunoglobulin diversity. In the
rabbit only a single germline VH gene is rearranged in the
majority of B‐cells; this then becomes a substrate for gene conversion by one
of the large number of VH pseudogenes. There are also large
numbers of VH pseudogenes and orphan genes (genes located
outside the gene locus, often on a completely different chromosome) in humans
that actually outnumber the functional genes, although there is no evidence to
date that these are used in gene conversion processes.
 |
Figure 4.14 Mutations
in regions of five IgM and five IgG monoclonal phosphorylcholine antibodies
generated during an antipneumococcal response in a single mouse are compared
with the primary structure of the T15 germline sequence. A line indicates
identity with the T15 prototype and an orange circle a single amino acid
difference. Mutations have only occurred in the IgG molecules and are seen in
both hypervariable and framework segments. (After Gearhart P.J. (1982) Immunology
Today 3, 107.) Although in some other studies somatic hypermutation
has been seen in IgM antibodies, the amount of mutation usually greatly
increases following class switching.