Pathogen Recognition Receptors Provide The First Line Of Detection For Microbial Antigen
As
we learned in Chapter 1, the innate immune system employs an impressive battery
of defense mechanisms that specifically detect the presence of invading
microbes, to coordinate a series of rapid responses that deal directly with the
invader, while at the same time sowing the seeds for a more specific and long‐lasting adaptive immune response. Over many millennia of co‐evolution,
vertebrate immune systems have become impressively adept at accurately
identifying the presence of potentially harmful microbes, through the detection
of microbial structures that are essential for viability and, therefore,
refractive to the pressures of natural selection. These conserved microbial
antigens, called pathogen‐associated molecular patterns (PAMPs),
are unique to individual classes of microbes, and as such, convey
pathogen‐specific information to the innate immune system, to facilitate an
appropriate response tailored to the particular threat at hand.
Detection
of PAMPs is facilitated by a family of evolutionarily conserved
germline‐encoded receptors called pathogen recognition receptors (PRRs),
expressed on innate immune cells such as DCs, macrophages, and neutrophils.
PAMP detection is often the first indication to the innate immune system of
microbial presence and consequently, PAMP‐induced PRR activation rapidly
promotes the production of a host of cytokines, chemokines, and type 1
interferons that mobilize innate immune cells to directly confront the invader.
Additionally, PRR stimulation acts as a crucial line of communication between
the innate and adaptive immune systems by instructing antigen‐presenting cells,
such as DCs, to effectively
license a
T‐cell‐mediated adaptive immune response against a particular
antigen. As will be discussed in later chapters, the particular mode of T‐cell
activation is further shaped by PRR‐ induced DC‐derived cytokines, which
effectively tailor the T‐ cell‐mediated response to the particular type of
microbe. As PRR signaling has also been shown to be important for instructing
B‐cells to respond to particular types of microbial antigen, it should be clear
that the recognition of microbial PAMP by PRRs plays a crucial role in
coordinating both innate and adaptive immune responses to infection.
To
date, several different classes of PRRs have been charac terized, including
Toll‐like receptors (TLRs), NOD‐like receptors (NLRs), RIG‐1‐like receptors
(RLRs), DNA receptors, and C‐type lectin‐like receptors, which together sense a
wide range of conserved microbial antigen. TLRs are among the best‐characterized
PRRs and we will next turn our attention to this important immune receptor
family.
Toll‐like receptors detect
a wide range of conserved microbial PAMP
Named
after a Drosophila protein that was originally discovered as important
for embryogenesis and later, as required for antifungal immunity, Toll‐like
receptors (TLRs) are a key family of mammalian PRRs
involved in the detection of a wide variety of PAMPs. To date, 10 TLRs have
been described in humans, and 12 have been characterized in mice. TLR1, 2, 4,
5, and 6 are expressed on the cell surface and detect ligands
from bacteria, fungi, protozoa, and certain self antigens, whereas expression
of TLR3, 7, 8, and 9 are confined to intracellular endocytotic
compartments, where they recognize nucleic acids signatures unique to
bacteria and viruses (Figure 4.30a).
TLRs
are type 1 integral membrane receptors composed of an extracellular
ligand‐binding domain, a single transmembrane helix, and an intracellular Toll/IL‐1R
(TIR) signaling domain, named because of
its homology to the signaling domains of the interleukin‐1 receptor
superfamily. Ligand binding induces dimerization of extracellular TLR domains,
which in turn facilitates the localization and subsequent dimerization of
intracellular TIR domains required for signaling. Dimerized TIR domains then
recruit various adaptors, including myeloid differentiation primary
response protein 88 (MyD88) (Figure 4.30b) and TIR‐domain‐containing
adaptor inducing interferon‐β (TRIF), which ultimately
promote activation of transcription factors such as nuclear factor kB (NFκB)
and interferon regulatory factors (IRFs), responsible for inducing expression
of cytokines, chemokines, and antimicrobial factors.
TLRs
belong to the leucine‐rich repeat (LRR) family of
proteins, with extracellular domains characterized by tandem repeats of LRR
modules of 20–30 amino acids in length, with the hydrophobic leucines spaced at
defined intervals. The leucines face toward the interior of the protein,
forming a hydrophobic core that acts to stabilize overall protein structure,
with variable regions facing outward to form a β arrangement gives TLRs a
classical solenoid‐like shape, with each LRR module organized
into adjacent, coiled, circular structures, similar to the way nuclear DNA is
wound around histones, while the β‐sheet of one LRR is arranged in parallel
with the β‐sheet of an adjacent LRR. As the β‐sheets are more tightly packed
than the rest of the LRR, the overall structure of the receptor is forced to
bend into a horseshoe shape, with β‐ sheets arranged on the
concave side (Figure 4.31a). Although the majority of LRR family proteins
interact with protein ligands, TLRs are distinct in their interaction with
nonprotein antigens, with ligands interacting at the concave or lateral sides
of the receptor.
Although
all TLRs share similar overall structure, they display considerable divergence
in their ligand‐binding affinities, driven mainly by differences in the size
and charge of ligand‐binding pockets, and their ability to engage in ligand‐induced
homodimerization (TLR3, TLR7) and ligand‐driven heter-
odimerization with other members of the TLR family (TLR2/1, TLR2/6),
and with non‐TLR co‐receptors (TLR4/ MD‐2) (Figure 4.30a). Regardless of the
ligand specificity of individual TLRs, ligand‐induced dimerization of adjacent
receptors results in a characteristic “m‐shaped” conformation, with
the TLRs interacting at their C‐termini to drive dimeriza tion of
intracellular TIR domains. To look at the structure of TLRs more closely, we
will next turn to possibly the best‐char acterized of these receptors, TLR4.
The TLR4/MD‐2 complex
detects microbial lipopolysaccharide
Lipopolysaccharide (LPS)
is an essential component of Gram‐negative bacterial cell walls, capable
of inducing potent immune responses at extremely low concentrations, which, if
left unchecked, can lead to septic shock and death. Such an acute response
suggests that mammalian innate immune systems have evolved to detect this PAMP
with exquisite sensitivity and this detection is carried out by TLR4, in
conjunction with its co‐receptor MD‐2, both of which are
abundantly expressed on the majority of innate immune cells, and on B‐ cells,
and barrier tissues at the front line of infection. This double team forms a 1
: 1 heterodimer, with TLR4‐bound MD‐2 acting as the primary binding
interface with LPS. Interaction between LPS and MD‐2 opens up MD‐2 residues
that promote stable interaction with adjacent TLR4 molecules, promoting
dimerization of adjacent TLR4/MD‐2 complexes, with the subsequent dimerization of intracellular TIR
domains that triggers signaling.
Native LPS is buried in bacterial
cell walls in a difficult‐to‐detect conformation, but is efficiently extracted
by a serum factor called LPS‐binding protein (LBP)
and facilitated by complement factors that punch holes in bacterial cell walls,
dispersing bite‐sized chunks of LPS‐containing material into the bloodstream.
LBP transfers LPS oligomers to CD14, which further splits them
into monomers, for presentation to the TLR4/MD‐2 complex for efficient
detection. Prior to LPS binding, TLR4 and MD‐2 are bound together as
heterodimers, with the 21 LRR TLR4 ectodomain arranged in the typical horseshoe
shape, and the smaller MD‐2 molecules bound to the lateral side, suspended
downwards in a hanging, flower basket‐like arrangement (Figure 4.31a). MD‐2 is
the main interactor with LPS and adopts a cup‐like structure, with two
antiparallel β‐sheets forming a stable barrel‐shaped core that can accommodate
lipid molecules of a defined size. LPS is a glycolipid with a hydrophobic lipid
A region attached to a carbohydrate chain and the number of lipid
chains in the lipid A segment appears to be a critical determinant of TLR4/MD‐2
complex activation, with six lipid chains forming the ideal number. Indeed, the
lipid A region is responsible for the majority of inflammatory activity of LPS,
with five lipid chains exhibiting 100‐fold lower activity and four lipid
chains, such as eritoan, acting as inhibitors. The crystal structure of the
TLR4 ectodomain/MD‐2/LPS complex illustrates the preference for six chains.
Five lipid A chains of LPS are buried deeply in the hydrophobic β‐pocket
of MD‐2, while the sixth lipid A residue is exposed, with negatively
charged phosphate groups making critical contacts with positively charged
residues on both MD‐2 and the TLR4 ectodomain. Importantly, these interactions
re‐orientate MD‐2 such that its F126 and L87 loops become
exposed and are now free to make contact with a separate, adjacent, TLR4
molecule, also bound to its own MD‐2, which, in turn, makes a reciprocal
interaction. This site of interaction between adjacent LPS and MD‐2 molecules
is called the dimerization interface and promotes dimerization of
adjacent TLR4/MD‐2 molecules with the resulting heterotetrameric complex of
TLR4–MD‐2–LPS, in a 2 : 2 : 2 ratio (Figure 4.31a). The net result of all these
interactions results in stable interaction between the C‐termini of two TLR4
ectodomains, forming an m‐shaped structure that facilitates close interaction
and subsequent dimerization of intracellular TIR domains (Figure 4.31a,b).
As noted above, TIR domain
dimerization is required for the recruitment of the TIR domain‐containing
adaptor MyD88, which recruits IRAK4 and IRAK2 in a defined structure that has
been dubbed the Myddosome, which relays the inflammatory signal
into the cell. We will look more closely at how the structure of the Myddosome
is organized to perform this task but first let us take a look at a TLR with
binding properties that are different from those of the TLR4/ MD‐2 complex,
TLR2.
TLR2/1/6
TLR2 plays a crucial role in the
recognition of microbial lipo- peptides, and mice deficient in this
receptor are at increased risk of infection with a variety of bacteria,
including S. pneumoniae and M. tuberculosis. Bacterial
lipoproteins are composed of a glycerol backbone with either two or three
attached acyl (fatty acid) chains. Gram‐negative bacteria possess
tri- acylated lipoproteins with two fatty acid chains, attached
by ester bonds to an N‐terminal cysteine, with the third lipid chain connected
to the cysteine by an amide bond, whereas lipoproteins from Gram‐positive
bacteria and mycoplasma are diacylated as they lack the amide‐bound
lipid chain and thus have just two fatty acid chains. Early gene knockout
studies showed that macrophages from TLR2‐deficient mice lost the ability to
respond to both di‐and triacylated lipoproteins from a variety of bacteria.
Interestingly, TLR1‐deficient macrophages lost the ability to respond to
triacylated lipoproteins only, whereas macrophages deficient in TLR6 failed to
respond to the diacylated form. These results strongly suggeste that TLR2
worked in conjunction with TLR1 to detect triacylated lipoproteins from
Gram‐positive bacteria, while it paired up with TLR6 for detection of
Gram‐positive bacteria bearing diacylated lipoproteins. Indeed, subsequent
crystal structures confirmed this data, showing that triacylated lipoproteins
simultaneously bound both TLR1 and TLR2, effectively acting as a bridge to draw
the two receptors close enough together for dimerization to occur, while
diacylated lipoproteins formed a complex with both TLR2 and TLR6.
Although TLR2 can directly bind
both di‐ and triglycerides without the need for intervention from TLR1 or TLR6,
this binding does not promote an optimal interaction between individual
lipoprotein‐bound TLR2 receptors and thus, the dimerization of adjacent TLR2
ectodomains required for intra cellular signaling fails to occur. This is due
to the fact that TLR2 efficiently binds the first two lipid chains on a
lipoprotein, leaving the rest of the molecule free to undergo specific
interactions with TLR1, in the case of the triacylated form, or TLR6 for
diaceylated lipoproteins. Indeed it is the specificity of TLR1 for triacylated
lipoproteins and TLR6 for diacylated lipoproteins that confers specificity on
the TLR2/1 and TLR2/6 complexes.
The ectodomains of all three TLRs
display the characteristic TLR horseshoe shape, with 20 LRR modules each
containing 24 residues, and can be divided into three distinct subdomains:
N‐terminal, central, and C‐terminal (Figure 4.32a). Although the N‐terminal
domain shares homology with other LRRs, the central and C‐terminal domains of
TLR1 and TLR2 deviate from the norm, with the border between these two domains
molded into ligand‐binding pockets, lined with hydrophobic residues.
The ligand‐binding pocket on TLR2 is large enough to accommodate the first two
fatty acid chains of a triacylated lipoprotein, while the third acyl chain fits
into a similar but smaller pocket on TLR1. The bound triacylated ligand now
effectively acts as a bridge to pull both TLRs close together,
allowing hydrophobic residues that surround the binding pockets on both TLRs to
form hydrogen bonds that further stabilize the interaction, pulling both TLRs
closer together (Figure 4.32). These ligand–TLR and TLR–TLR interactions result
in dimerization of TLR1 and TLR2 at their C‐termini, forming the distinctive “m
shape” that facilitates localization of intracellular TIR domains.
Although TLR1/2 complexes
efficiently bind triacylated lipoproteins, why are TLR2/6 complexes specific
for diacylated ligands? The answer lies in a number of important structural
differences between TLR1 and TLR6 in their ligand‐binding and dimerization
surfaces. Whereas TLR1 can accommodate an acyl chain in its C‐terminal
ligand‐binding pocket, this pocket in TLR6 is partially blocked by the
bulky side chains of two phenylalanine residues, reducing the pocket
size by half and restricting ligand entry. Indeed, mutation of this region of
TLR6 to mimic that found in TLR1 allows TLR6 to efficiently bind triacylated
ligands, underlying the importance of these C‐terminal phenylalanine residues
in conferring specificity for diacylated lipoproteins. Although TLR6 lacks a
ligand‐binding pocket that could accommodate an acyl chain, it makes up for it
in a superior ability to bind the peptide part of diacylated lipoproteins. As
in the TLR1/2 complex, the two acyl chains of the lipopeptide are buried in the
C‐terminal pocket of TLR2, while the exposed peptide region of the ligand forms
a number of strong hydrogen bonds with both TLR2 and TLR6 (Figure 4.32). In addition,
an extensive region on TLR6 also makes direct contact with TLR2, forming stable
hydrogen bonds that account for an increase in protein–protein
inter-action of at least 80% when compared with TLR1/2. These
interactions combine to drive TLR2 and TLR6 close enough together for
dimerization and intracellular signaling to occur.
Although we have focused on the
extracellular TLR domain interactions that are brought about by ligand binding,
the associated re‐orientation of intracellular domains required to drive
signaling is equally as important and it is to this that we will next turn our
attention.
Dynamic structural rearrangements propagate intracellular TLR
signaling
Regardless of the nature of
ligand‐induced dimerization of individual TLR ectodomains, dimerization at the
C‐termini re‐orientates the receptors such that the intracellular TIR
domains colocalize and undergo the dimerization required to recruit TIR
domain‐containing adaptors. Interestingly, extensive artificial truncation of
TLR ectodomains triggers receptor auto‐activation, which suggests that in their
unbound forms, the ectodomains may act to inhibit an intrinsic tendency for the
transmembrane and intracellular domains to dimerize.
There are five TIR
domain‐containing adapters that transmit TLR signals into the cell, with MyD88
required at a proximal level for the signaling of all TLRs except TLR3,
which uses TRIF exclusively. In the case of TLR4, ligand binding
promotes ectodomain dimerization, allowing the TIR domains to dimerize and
recruit six molecules of MyD88, in conjunction with the bridging molecule MyD88
adaptor‐like protein (MAL). Close contact between the
death domains of MyD88 is then thought to facilitate recruitment of four
molecules of the death domain‐containing adaptor IRAK4, which in
turn, recruits four molecules of IRAK2, forming a higher order,
column‐like structure that has been dubbed the Myddosome, which
is responsible for activating NFκB.
TIR domain structure can be
subdivided into a central β‐ sheet, organized into four or five parallel
β‐strands (the βA–βE strands), with five α‐helices (αA–αE helices), connected
to the edges of the sheet by a series of loops. Some of these loops play a critical
role in signal transduction, such as the BB loop that joins the
βB strand of the β‐sheet with the αB α‐helix. A polymorphism in this region in
TLR4 in the CHC3H/HeJ strain of laboratory mice completely kills signaling from
the receptor and renders these mice incapable of responding to LPS. Although
dimerized TIR domains have proved difficult to crystallize, mutational and
inhibitor studies have shed light on the method of TIR domain dimerization,
with the BB loop of adjacent TIRs predicted to form an extensive interface. In
addition, regions within the BB loop also make direct contact with the TIR
domain of MAL, which acts as a bridging molecule to stabilize TLR4–MyD88
interaction.
TLR4 can also signal through the
TIR domain‐containing adaptor TRIF, in conjunction with the
bridging molecule TRIF‐related adaptor molecule (TRAM), to drive
activation of IRF3 and expression of interferon genes. TRAM is recruited to
TLR4 only after receptor endocytosis, suggesting that a possible conformational
change in the receptor, driven by the acidic environment of the endosome, may
be required for TRAM binding and subsequent TRIF recruitment. Interestingly,
the TIR domain of TLR3, which signals exclusively through TRIF, contains an
alanine in the BB loop, rather than a proline like all the other TLRs, and
mutation of this residue in TLR3 to proline changes specificity of TLR3 from
TRIF to MAL/ MyD88, with associated NFκB signaling as opposed to IRF‐ dependent
events.
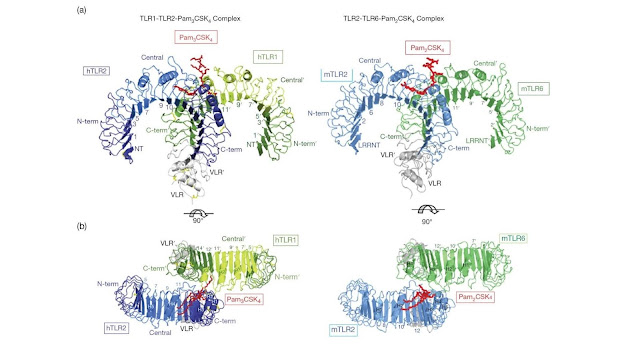
Figure 4.32 Overall structure of the human TLR1–TLR2–Pam3CSK4
complex and the mouse TLR2–TLR6–Pam2CSK4 complex. To facilitate crystallization
and structure determination the LRR C‐terminal and the last one or two LRRs of
TLRs 1, 2, and 6 were replaced by corresponding regions of a hagfish VLR. The
TLR1, TLR2, TLR6, and VLR fragments in the TLR–VLR hybrids are shown
schematically in green (TLR1 and TLR6), blue (TLR2), and gray (VLR). Pam3CSK4 and
Pam2CSK4 are shown in red. Some LRR modules are numbered and the N‐terminal,
central, and C‐terminal subdomains are labeled. (a) Side view, (b) top view.
(Source: Jin M.S. et al. (2007)
Cell 130, 1071–1082 and Kang
J.Y. et al. (2009) Immunity 31, 873–884.
Reproduced with permission of Nature Publishing Group.)
MyD88 and TRIF form higher order complexes
In addition to a TIR domain,
MyD88 also contains a death domain (DD), common in
proteins associated with apoptosis as well as immunity. The MyD88 DD provides a
platform for recruitment of the DD‐containing IRAK4, which in turn recruits
IRAK2 via DD interactions. Death domains bestow on these proteins the ability
to form hetero‐oligomers and the crystal structure of the MyD88–IRAK4–IRAK2
complex has illuminated the impressively ordered nature of this signaling
platform (Figure 4.33). Six–eight molecules of MyD88 recruit four molecules of
IRAK4, which in turn recruit four molecules of IRAK2 in a helical,
three‐layered complex called the Myddosome, driven by DD–DD
interactions. The importance of this com plex for TLR signaling is illustrated
by a naturally occurring polymorphism in the DD of MyD88 that renders these
complexes defective for both signaling and Myddosome formation.
In contrast to MyD88, the larger
TRIF molecule lacks a DD, instead containing an α‐helical N‐terminal domain
(TRIF‐NTD) that is thought to autoinhibit activation of the resting TRIF
protein by obscuring the binding sites of downstream adaptors. Binding of TRIF
to TLR3 or TLR4/TRAM displaces the TRIF‐NTD and frees up a proline‐rich
region in the protein, which facilities recruitment of tumor necrosis
fac tor receptor‐associated factor 2 (TRAF3) and TANK‐binding kinase 1 (TBK1)
for activation of IRFs. In addition, the TRIF receptor‐interacting protein
(RIP) homotypic interaction motif (RHIM) is also liberated to recruit RIP
kinase 1, resulting in both FADD‐dependent apoptosis and NFκB activation.
Crystal structures of the TRIF complexes have not yet been resolved to answer
the question of whether or not they form higher order complexes like the
Myddosome, but the current thinking is that a similar TRIF‐containing complex
may be formed.
Figure 4.33 Myddosome structure. (a) Ribbon diagram of
Myddosome structure, with the six MyD88 molecules in cold colors, the four
IRAK4 molecules in earth‐tone colors, and the four IRAK2 molecules in warm
colors. (b) Surface diagram of the complex with each subunit labeled using the
same color coding as in (a). M, MyD88; I4, IRAK4; I2, IRAK2. (Source: Lin S.C. et al. (2010) Nature 465, 885–890. Reproduced with permission of Nature
Publishing Group.)
C‐type lectin‐like receptors detect fungal antigen
C‐type lectin‐like receptors (CLRs) form a large and varied family of receptors that
share in common a C‐type lectin‐like domain (CTLD)
and function in a variety of scenarios, from cell–cell adhesion to immune
signaling and apoptosis. Although the CTLD bears structural homology to the
carbohydrate‐ binding domains found in carbohydrate‐binding proteins, CTLDs are
more varied and are not necessarily restricted to carbohydrate ligands. This
family of receptors can be loosely subdivided by their requirement for calcium
for functional ligand binding and on the type of intracellular signaling domain
that can possess activating ITAMs or inactivating ITIMs.
Ligand recognition and signal transduction by activating CLRs is broadly
similar to the TLR scenario; ligand binding promotes receptor ectodomain
dimerization, which then dimerizes and activates the intracellular ITAM motifs
to recruit
ITAM‐containing adapter molecules
such as Syk kinase, to promote activation of proinflammatory transcription
factors such as NFκB.
Although many members of the CTLD
family bind a variety of carbohydrates from a number of different
microorganisms (e.g., dectin‐1 binds β‐glucan and dectin‐2 recognizes mannose)
other members, such as the lipid‐binding mincle, can also bind non carbohydrate
ligands. The fungal β‐glucan‐ binding receptor dectin‐1 is the
best‐characterized CTLD receptor and we will now look more closely at its mode
of action.
Dectin‐1 recognizes fungal β‐glucan
Immune responses to fungal
infections are mediated mainly by CTLD receptors, with the detection of
β‐glucans by dectin‐1 playing a particularly important role in antifungal
immunity. Mice deficient in this receptor display marked defects in immune cell
infiltration during fungal challenge and are highly susceptible to infection
with Candida albicans, while dectin‐1 also detects β‐glucans from a
range of other fungi, including Saccharomyces, Penicillium, and Aspergillus.
As highly conserved and essential components of the cell wall of certain fungi
and baker’s yeast, β‐glucans certainly fit the bill as classical PAMPs.
Dectin‐1 can recognize β‐1,3 and β‐1,6‐linked glucans from fungi,
plants, and bacteria, with the best‐characterized ligand, zymosan from yeast
cell walls, binding with high affinity. The expression of dectin‐1 on dendritic
cells, monocytes, macrophages, and neutrophils places it on the front line of
antifungal immunity, where receptor activation can trigger pathogen phagocytosis
or the generation of antifungal cytokines and chemokines.
With a single extracellular CTLD,
a transmembrane region and a cytoplasmic ITAM, ligand binding is thought to
promote dimerization of the dectin‐1 ectodomain, required to activate intracellular
ITAMs. Unlike other members of the CTLD receptor family, ligand binding occurs
in the absence of calcium. Crystal structure of the extracellular portion of
dectin‐1 illustrates that it adopts a similar conformation to other
CTLD‐containing receptors, with two antiparallel β‐sheets and two α‐helices,
with the N‐ and C‐termini in close proximity (Figure 4.34). Sequence analysis
has highlighted a number of surface hydrophobic residues that could play a role
in ligand binding, and mutational studies have identified two residues, Trp221
and His223 in the third ticularly important for ligand
recognition. Mutation of these residues to an alanine blocked the interaction
of β‐glucan with the receptor, while a dectin‐1 antibody that efficiently
inhibited β‐glucan binding failed to bind to the W221A mutant, suggesting the
region plays a key role in ligand interaction. This region adopts a shallow
hydrophobic groove in the crystal structure of dectin‐1, but no ligands were
observed binding in this pocket, possibly due to technical constraints in
achieving crystallization of β‐glucan ligands of sufficient size. Indeed,
cell‐based studies have suggested that the minimum size of β‐glucan sufficient
to bind the receptor is no smaller than 10‐mer, which could certainly be
accommodated in this groove. Although the current crystal structure is
inconclusive, it remains likely that β‐glucan binding acts to bridge adjacent
dectin‐1 molecules to facilitate ITAM dimerization and recruitment of Syk
kinase and, potentially, Raf, which can both drive immune signaling through
NFκB activation. Activated Syk also drives calcium‐dependent outcomes such as
NFAT activation, with associated cytokine secretion.