The acquired immune responses
mounted by lymphocytes depend upon specific recognition of antigen by the
B‐cell receptor (BCR, a transmembrane version of the antibody molecule) or the
T‐cell receptor (TCR). Following clonal selection the antigen‐specific
lymphocytes undergo proliferation to produce sufficient numbers of effector
cells and also to generate memory cells. In the case of B‐cells the main
effector cells are the plasma cells that secrete a soluble version of the same
antibody that was used as the BCR on the original B‐cell. In the case of
T‐cells the effector cells are cytokine‐secreting helper or regulatory cells,
or cell‐killing cytotoxic cells.
Introduction
In acquired immunity, specific
antigens are recognized by two classes of molecules: (i) antibodies,
present either as soluble proteins or as transmembrane molecules on the surface
of
B‐cells; and (ii) T‐cell
receptors, present as transmembrane molecules on the surface of T‐cells.
Antibodies recognize antigens on the outside of pathogens or as soluble
material such as toxins, whereas αβ T‐cell receptors recognize peptides in the
context of MHC molecules on the surface of host cells. Antibodies
can thus be thought of as scanning for foreign material directly whereas
T‐cells (particularly cytotoxic T‐cells) are scanning for cells that are
infected with pathogens.
Figure 5.1 Complementarity
of the antibody combining site and the epitope recognized on the antigen. The
structure of the complex of the Fab of the antibody pertuzumab and its antigen
HER2 is shown. HER2, the human epidermal growth factor receptor, is
overexpressed on some breast cancer cells and pertuzumab is an antibody,
similar to Herceptin®, with potential as a therapeutic against breast cancer.
Below, the two molecules are shown separately with the interaction footprint
shown on each. (Source: Robyn Stanfield. Reproduced with permission.)
What antibodies see
Antibodies recognize molecular
shapes (epitopes) on antigens. Generally, the better the fit of the epitope (in
terms of geometry and chemical character) to the antibody combining site, the
more favorable the interactions that will be formed between the antibody and
antigen and the higher the affinity of the antibody for antigen. The affinity
of the antibody for the antigen is one of the most important factors in
determining antibody efficacy in vivo.
Epitopes come in a huge variety
of different shapes, as do antibody combining sites. Protein surfaces are
typically recognized by a complementary surface in the antibody combining site,
as illustrated in Figure 5.1 that shows how an antibody recognizes an epitope
on the human epidermal growth factor receptor HER‐2. The extent of complementarity
of the inter- acting surfaces is readily appreciated.
The area of antigen that contacts
antibody is referred to as a footprint and is typically between about 4 and 10
nm2. Footprints are of somewhat different sizes and irregular shapes; a
projection of a 2.5 × 2.5 nm square onto a series of protein antigens gives an
appreciation of the approximate size of typical antibody footprints (Figure
5.2).
Antibodies recognize a
topographic surface of a protein antigen. Most usually, key residues in the
epitope will arise from widely different positions in the linear amino acid
sequence of the protein (Figure 5.3). This follows because of the manner in which
proteins are folded: the linear sequence typically snakes from one side of the
protein to the other a number of times. Such epitopes are described as discontinuous.
Occasionally, key residues arise from a linear amino acid sequence. In such
cases, the antibody may bind with relatively high affinity to a peptide
incorporating the appropriate linear sequence from the antigen. Furthermore,
the free peptide may inhibit the antigen binding to the antibody. The epitope
in such cases is described as continuous. An example of a
continuous epitope would be a loop on the surface of the protein for which an
antibody recognized successive residues in the loop. It should be noted,
however, that an antibody that recognizes a continuous epitope does not bind a
random or disordered structure. Rather it recognizes a defined structure that
is found in the complete protein but can readily be adopted by the shorter
peptide. The structure of an antibody that recognizes a linear epitope in
complex with a peptide that contains the epitope is shown in Figure 5.4; note
that the structure of the peptide is largely helical in this example.
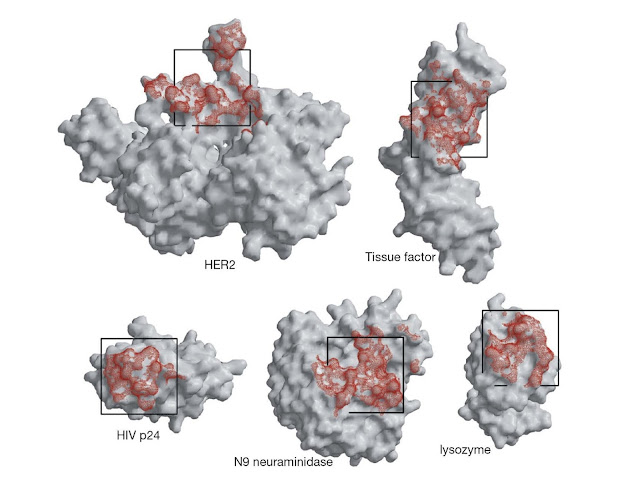 |
Figure 5.2 Antibody footprints (red) on a range of
antigens. These footprints are determined from crystal structures of the
antigens with antibody bound. The footprints are irregular but can be very
roughly represented as a square of dimensions 2.5 × 2.5 nm as shown. (Source:
Robyn Stanfield. Reproduced with permission.)
 |
Figure 5.3 Residues contributing to epitopes on the
folded peptide chain of myoglobin. Amino acid residues 34, 53, and 113 (black)
contribute to the binding of a monoclonal antibody (mAb) and residues 83, 144,
and 145 to the binding of another mAb (red). Both epitopes are clearly
discontinuous. By contrast, a third mAb binds to residues 18–22 (green). The
mAb binds to isolated peptides containing the sequence corresponding to
residues 18–22. The epitope is described as continuous. Much of the myoglobin
structure is in α‐helical conformation. (Source: Adapted from Benjamin D.C. et al. (1986) Annual Review of Immunology 2, 67.)
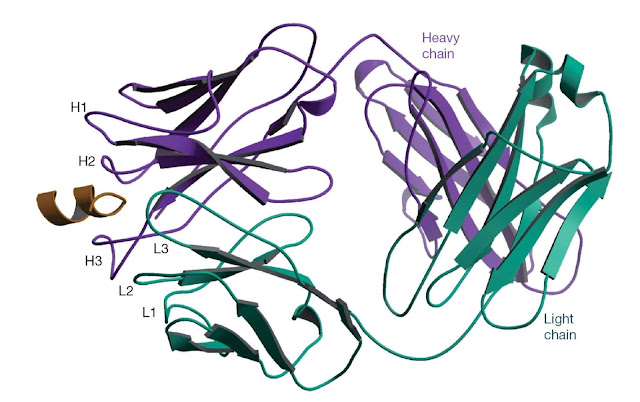 |
Figure 5.4 The structure of an antibody bound to a
peptide corresponding to a linear epitope. The antibody 4E10 neutralizes HIV by
binding to a linear epitope on the glycoprotein gp41 on the surface of the
virus. The antibody binds to peptides containing the amino acid sequence NWFDIT
and peptides containing this sequence can inhibit the binding of 4E10 to gp41.
The structure of the Fab fragment of 4E10 bound to a peptide (gold) containing
the NWFDIT sequence shows the peptide adopts a helical conformation. It is
likely that the antibody recognizes its epitope in a helical conformation on
the virus. (Source: Rosa Cardoso. Reproduced with permission.)
The antibody complementarity determining regions (CDRs) contact the
epitope
The antibody combining site can
vary greatly in shape and character depending upon the length and
characteristics of the CDRs. Generally most or all of the CDRs (although by no
means all of the residues making up a CDR) contribute to anti-gen binding but
their relative contributions vary. The heavy chain CDRs, and particularly CDR
H3, tend to contribute disproportionately more to antigen binding. The CDR H3
in human antibodies can be quite long and has a finger‐like appearance that
could be used to bind into cavities on the anti-gen. The combining site of
antibodies against smaller molecules such as carbohydrates and organic groups
(haptens) are often more obviously grooves or pockets rather than the extended
surfaces typically found in antiprotein antibodies. It should also be noted
that framework region (FR) residues can also contribute to antigen binding. For
highly somatically mutated antibodies, such as those to HIV, quite extensive
contacts between FR residues and the viral surface antigen are observed.
Structural changes and
conformational rearrangements can occur in antibodies or antigens on
interaction. In other words, on some occasions, the relationship between
antibody and antigen will be like a “lock and key” but on other occasions the
lock or key or both can be deformed to make a good fit. For the antibody,
possible conformational changes include side‐chain rearrangements, segmental
movements of CDRs or of the main‐chain backbone, and rotation of the VL
–VH domain upon antigen binding. Large changes in the conformation
of the CDR H3 have been documented in crystal structures of Fab complexes. As
shown in Figure 5.5, an antibody to progesterone has a very hydrophobic
combining pocket, which is normally filled with a tryptophan from the CDR H3.
Antigen binding involves this residue moving out of the pocket, the antigen
molecule moving in and the trytophan stabilizing the antigen binding. As more
and more structures have been solved it has become clear that antibody antigen interactions
come in all shapes and sizes with few general rules. It is important to bear in
mind that high‐affinity antibodies evolve in each individual following rounds of
mutation and selection. There are multiple ways in which high‐affinity
recognition of an antigen can be achieved, and indeed no two antibody antigen
interactions are exactly the same.
Figure 5.5 Conformational change in an antibody
combining site. (a) An anti‐progesterone antibody has a very hydrophobic pocket
that is filled by a tryptophan residue (colored red) in the free antibody. (b)
To bind progesterone (dark blue), the tryptophan residue swings out of the
pocket and the antigen gains access. (Source: Robyn Stanfield. Reproduced with
permission.)
Antigens versus immunogens
An epitope on an antigen may bind
very tightly to a given anti-body but it may elicit such antibodies
infrequently when the antigen is used to immunize an animal. In other words,
there may be a perfectly good site on a pathogen for antibody binding but the
antibody response to that site is so poor it cannot contribute to antibody
protection against the pathogen. We say that the site has low immunogenicity
and the consequences can clearly be great.
An extreme example of the
distinction between the ability to be recognized by an antibody (which we will
term antigenicity) and the ability to elicit antibodies when used to immunize
an animal (which we will term immunogenicity) is provided by experiments using
small molecules known as haptens such as m‐aminobenzene sulfonate.
Immunization with free hapten produces no antibodies to the hapten (Figure
5.6). However immunization with hapten groups linked to a protein carrier
generates antibodies that react with high affinity to hapten alone or linked to
a molecule other than the carrier. It is logical to refer to the hapten as the
antigen and the hapten protein complex as the immunogen, although strictly the
word “anti-gen” is derived from “antibody generating” substance.
 |
Figure 5.6 Antigenicity and immunogenicity. A free small
molecule hapten will not induce antibodies if injected in to an animal.
However, high‐affinity antibodies specific for the free hapten can be obtained
by injecting the hapten conjugated to a protein carrier molecule such as
ovalbumin.






