The IgG Molecule
In IgG, the
Fab arms are linked to the Fc by an extended region of polypeptide chain known
as the hinge. This region tends to be exposed and sensitive to attack by
proteases that cleave the molecule in to its distinct functional units arranged
around the four‐chain structure (Milestone 3.1). This structure is repre
sented in greater detail in Figure 3.2a. The light chains exist in two forms,
known as kappa (k) and lambda (λ). In humans, k chains are somewhat more
prevalent than λ; in mice, λ chains are rare. The heavy chains can also be
grouped into different forms or subclasses, the number depending upon the
species under consideration. In humans there are four subclasses hav
ing heavy chains labeled γ1, γ2, γ3, and γ4, which give rise to the IgG1, IgG2,
IgG3, and IgG4 subclasses. In mice, there are again four subclasses denoted
IgG1, IgG2a, IgG2b, and IgG3. The subclasses – particularly in humans – have
very similar primary sequences, the greatest differences being observed in the
hinge region. The existence of subclasses is an important feature as they show
marked differences in their ability to trigger effector functions. In a single
molecule, the two heavy chains are generally identical, as are the two light
chains. The exception to the rule is provided by human IgG4, which can exchange
heavy–light pairs between IgG4 molecules to pro duce hybrids. As the Fc parts
of the exchanging molecules are identical, the net effect is Fab arm exchange
to generate IgG4 antibodies having two distinct Fab arms and dual specificity.
The amino
acid sequences of heavy and light chains of antibodies have revealed much
about their structure and function. However, obtaining the sequences of
antibodies is much more challenging than for many other proteins because the
population of antibodies in an individual is so incredibly heterogeneous. The
opportunity to do this first came from the study of myeloma proteins.
In the human disease known as multiple myeloma, one cell making one particular
individual antibody divides over and over again in the uncontrolled way a
cancer cell does, without regard for the overall requirement of the host. The
patient then possesses enormous numbers of identical cells derived as a clone
from the original cell and they all synthesize the same immunoglobulin – the
myeloma protein – which appears in the serum, sometimes in very high
concentrations. By purification of myeloma proteins, preparations of a single
antibody for sequencing and many other applications can be obtained. An
alternative route to single or monoclonal antibodies arrived with
the development of hybridoma technology. Here, fusing individual
antibody‐forming cells with a B‐cell tumor produces a constantly dividing clone
of cells dedicated to making the one antibody. Finally, recombinant
antibody technologies, developed most recently, provide an excellent
source of monoclonal antibodies.
Sequence
comparison of monoclonal IgG proteins indicates that the carboxy‐terminal
(C‐terminal) half of the light chain and roughly three‐quarters of the heavy
chain, again C‐terminal, show little sequence variation between different IgG
molecules. By contrast, the amino‐terminal (N‐terminal) regions of about 100
amino acid residues show considerable sequence variability in both chains.
Within these variable regions there are relatively short sequences that show
extreme variation and are designated hypervariable regions. There are three of
these regions or “hot spots” on the light chain and three on the heavy chain.
As the different IgGs in the comparison recognize different antigens, these hypervariable
regions are expected to be associated with antigen recognition and
indeed are often referred to as complementarity determining regions (CDRs).
The structural setting for the involvement of the hypervariable regions in
antigen recognition and the genetic origins of the constant and variable
regions will be discussed shortly.
The
comparison of immunoglobulin sequences also reveals the organization of IgG
into 12 homology regions or domains, each possessing an internal
disulfide bond. The basic domain structure is central to an understanding of
the relation between structure and function in the antibody molecule and will
be taken up shortly. However, the structure in outline form is shown in Figure
3.2b,c. It can be seen that the light chain consists of two domains, one
corresponding to the variable sequence region discussed earlier and designated
the VL (variable light) domain and the other corresponding to a constant region
and designated the CL (constant light) domain. The IgG heavy chain consists of
four domains, the VH and CH1 domains of the Fab arms
being joined to the CH2 and CH3 domains of Fc via the
hinge. Antigen binding occurs at the tips of the Fab arms and involves the VL
and VH domains. Effector molecule binding occurs at the Fc stem and
involves the CH2 and/or CH3 domains.
It is also
clear (Figure 3.2b,c) that all of the domains except for CH2 are in
close lateral or “sideways” association with another domain: a phenomenon
described as domain pairing. The CH2 domains have two sugar chains
interposed between them. The domains also exhibit weaker cis interactions
with neighboring domains on the same polypeptide chain.
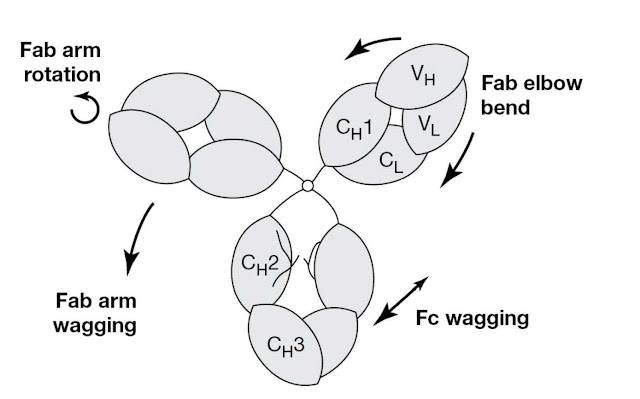
Human IgG1
is shown in Figure 3.2 as a Y‐shaped conformation with the Fab arms roughly in
the same plane as the Fc. This is the classical view of the antibody molecule
that has adorned countless meetings ads and appears in many company logos. In
reality, this is likely just one of many shapes that the IgG molecule can adopt
as it is very flexible, as illustrated in Figure 3.3. It is
believed that this flexibility may help IgG function. Thus Fab–Fab flexibility
gives the antibody a “variable reach,” allowing it to grasp antigenic
determinants of different spacings on a foreign cell surface or to form
intricate immune complexes with a toxin (imagine a Y to T shape change). Fc–Fab
flexibility may help antibodies in different environments, on foreign cells for
example, to interact productively with common effector molecules. Figure 3.4
shows the complete structure of a human IgG1 antibody molecule determined by
crystallography. The structure is quite removed from the classical symmetrical
Y shape. The Fc is closer to one Fab arm than another and is rotated relative
to the Fab arms. This is simply a “snapshot” of one of the many conformations
that the anti body can adopt by virtue of its flexibility.
The
structural organization of IgG into domains is clearly evident from Figure
3.2–Figure 3.4. Each of these domains has a common pattern of polypeptide chain
folding (Figure 3.5). This pattern, the “immunoglobulin fold,” consists of two
twisted stacked β‐sheets enclosing an internal volume of tightly packed
hydrophobic residues. The arrangement is stabilized by an internal disulfide
bond linking the two sheets in a central position (this internal bond is seen
in Figure 3.2a). In a constant type Ig domain, one sheet has four and the other
three anti‐parallel β‐strands. These strands are joined by bends or loops that
generally show little secondary structure. Residues involved in the β‐sheets
tend to be conserved while there is a greater diversity of residues in the
loops. The chain folding illustrated in Figure 3.5 is for a constant domain.
The β‐sheets of the variable domain are more distorted than those of the co
stant domain and the variable domain possesses an extra loop.
Structure of Fab fragment
The Fab
fragment pairs VH and VL domains and CH1 and CL
domains (Figure 3.6). The VH and VL domains are paired by
contact between the two respective three‐strand β‐sheet layers (red in Figure
3.5) whereas the CH1 and CL domains are paired via the
two four‐strand layers (blue in Figure 3.5). The interacting faces of the
domains are predominantly hydrophobic and the driving force for domain pairing
is thus the removal of these residues from the aqueous environment. The
arrangement is further stabilized by a disulfide bond between CH1
and CL domains.
In contrast
to the “sideways” interactions, the “longwise” or cis interactions
between VH and CH1 domains and between VL and CL
domains are very limited and allow bending about the “elbows” between these
domains. Elbow angles seen in crystal structures vary between about 117° and
249°.
The antibody combining site
Comparison
of antibody sequence and structural data shows how antibodies are able to
recognize an enormously diverse range of molecules. Sequence data show that the
variable domains have six hypervariable regions that display great variation in
amino acids between different antibody molecules (Figure 3.7). Structural data
of antibody antigen complexes reveal that these hypervariable regions, or
complementarity determining regions, come together in 3D space to form the
antigen‐binding site, often also termed the antibody combining site (Figure
3.8).
Structure of Fc
For the Fc
of IgG (Figure 3.9), the two CH3 domains are classically paired, whereas the
two CH2 domains show no close interaction, but have interposed
between them two branched N‐linked carbohydrate chains that have limited
contact with
one another.
The carbohydrate chains are very heterogeneous. The CH2 domains
contain the binding sites for several important effector molecules, complement
C1q and Fc receptors in particular, as shown. The neonatal Fc receptor, which
is important in binding to IgG and maintaining its long half‐life in serum,
binds to a site formed between CH2 and CH3 domains.
Protein A, much used in purifying IgGs, also binds to this site.
The hinge region and IgG subclasses
The term “hinge”
arose from electron micrographs of rabbit IgG, which showed Fab arms assuming
different angles rela tive to one another from nearly 0° (acute Y‐shaped) to
180° (T‐shaped). The Fab was specific for a small chemical group, dinitrophenyl
(DNP), that could be attached to either end of a hydrocarbon chain. As shown in
Figure 3.10 and
Figure 3.11,
different shapes were observed as the Fab arms linked together the bivalent
antigen molecule using different Fab–Fab arm angles. Other biophysical
techniques have demonstrated hinge flexibility in solution. The function of
this flexibility has generally been seen as allowing divalent recognition of
variably spaced antigenic determinants. The IgG class of antibody in humans
exists as four subclasses and the biggest difference between the subclasses is in
the nature and length of the hinge. IgG1 has been shown above. IgG3 has a hinge
that, if fully extended, would be about twice the length of the Fc, thereby
potentially placing the Fab arms far removed from the Fc. In contrast, IgG2 and
IgG4 have short, compact hinges that probably lead to close approach of Fab and
Fc. Interestingly, IgG1 and IgG3 are generally superior at mediating effector
functions such as complement activation and ADCC relative to IgG2 and IgG4.